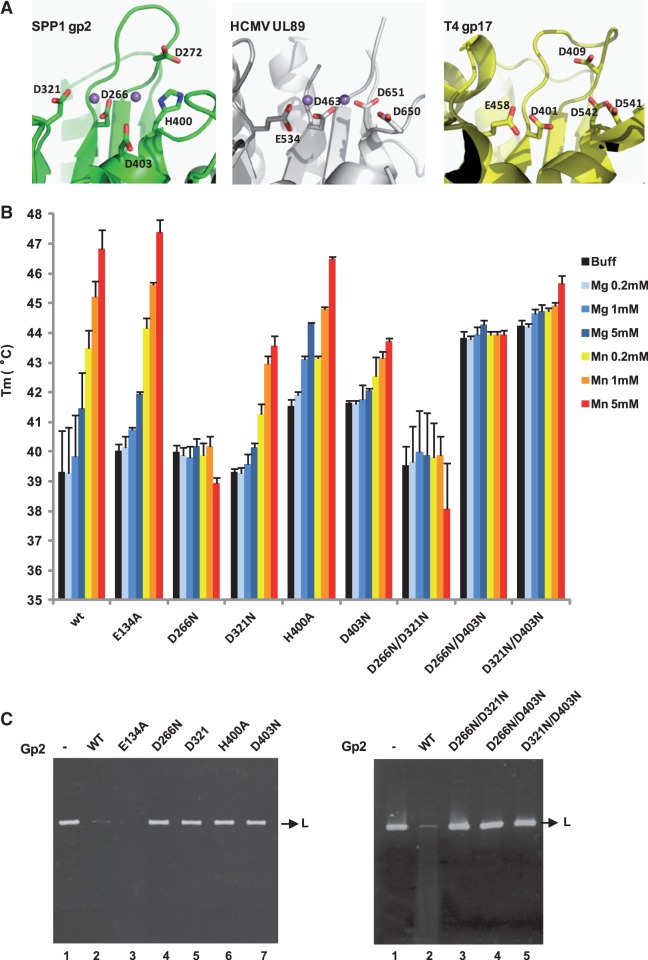
Figure 5.
Thermal stability assays and in vitro nuclease activity of gp2 mutant proteins. (A) Conservation of the terminase nuclease catalytic site of phage SPP1 (gp2; PDB accession code 2WC9), of HCMV (cytomegalovirus) (UL89; PDB 3N4Q) and of phage T4 (gp17; PDB 3CPE). The side chains of residues involved in metal coordination/catalysis are shown as sticks. The Mn2+ ions (magenta spheres) in SPP1 gp2 and in HCMV UL89 structures are 4 and 3.6 Å far apart, respectively. The structures were aligned with DALI and the figure prepared with PyMol. (B) Thermal stability of wild-type (wt) and gp2 mutant proteins analyzed as in Figure 4 in the absence of divalent metal ion (black bars) or in the presence of different concentrations (200 µM, 1 or 5 mM) of Mg2+ (light to dark blue bars) or Mn2+ (orange to red bars). The Tm values (°C) shown are the mean ± SD of results from at least three independent experiments. All proteins showed a single-transition denaturation curve. (C) In vitro nuclease activity of gp2 mutants. The nuclease activity of the gp2 single (left panel) and double (right panel) mutants was checked in an in vitro nuclease assay, as described under ‘Materials and Methods’ section, using a linear plasmid DNA substrate in the presence of 2.5 mM Mn2+.