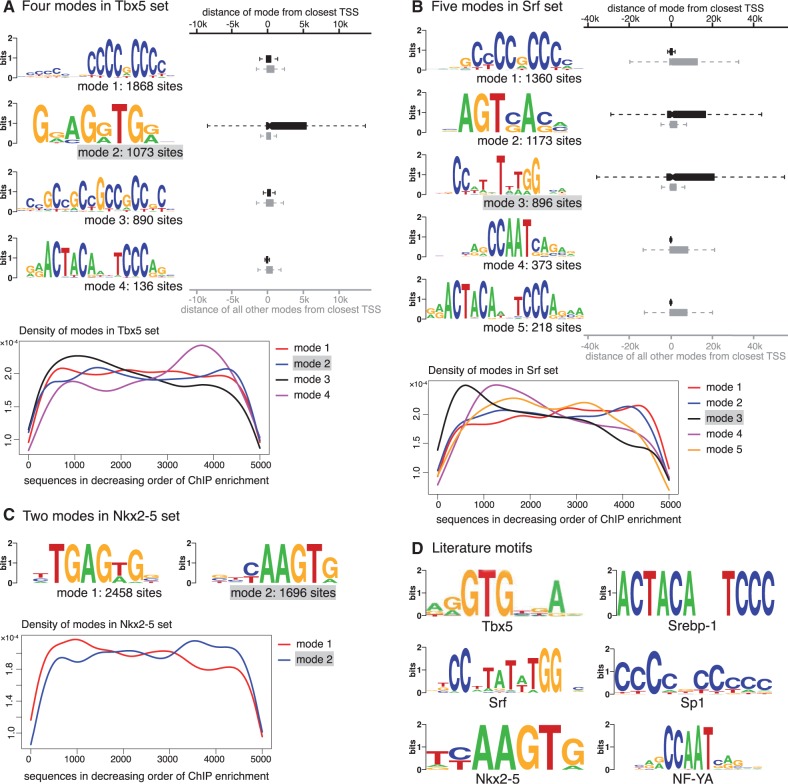
Figure 3.
Modes in three HL1 ChIP-Seq datasets. (A) Four modes are identified in Tbx5-bound sequences: motif for mode 2 matches the literature consensus. The distances to the closest TSS of the sequences in this mode (red boxplot) are significantly larger than sequences in other modes (blue boxplot). The density plot at the bottom illustrates the presence of each mode across sequences sorted by decreasing ChIP enrichment score. (B) Five modes are identified in Srf-bound sequences: motif for mode 3 matches literature consensus. The distances to the closest TSS of the sequences in this mode (red boxplot) are significantly larger than sequences in other modes (blue boxplot). The same holds true also for mode 2. Other three modes are enriched near the TSS. The density plot at the bottom shows that mode 3, which matches the Srf literature consensus, is more prevalent in the top 1000 sequences. (C) Two modes are identified in the Nkx2-5-bound sequences: motif for mode 2 matches literature consensus. The density plot indicates that this mode is slightly more prevalent in the lower half of the sequences. (D) Literature motifs for Tbx5 (reproduced from 31), Srebp1 (32), Srf (33), Sp1 (34), Nkx2-5 (33) and NF-YA (34).