Figure 5.
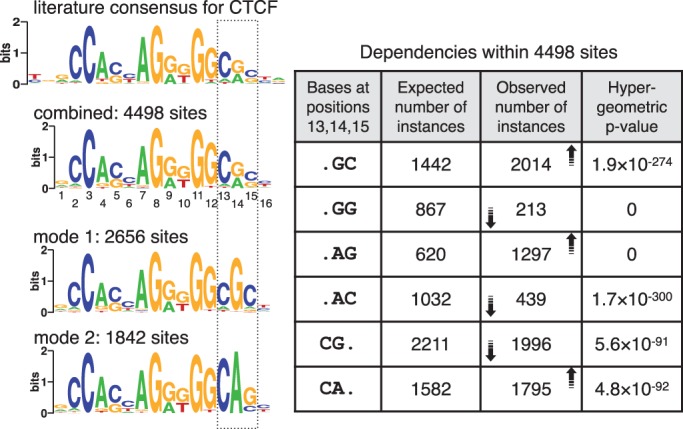
Two modes identified in cohesin-bound sequences in mouse ES cells. Conventional motif discovery programs return a motif similar to the CTCF motif shown on top. MuMoD identifies two modes, whose combination also resembles the CTCF motif. The table on the right describes dependencies within positions 13–15 in the combined motif. The first column considers different dinucleotide combinations; the second column shows the expected number of instances of each combination, assuming that the combined PSSM is an appropriate representation of the binding sites; the third column shows the number of instances actually observed, with the arrows indicating the increase or decrease in numbers with respect to the expected instances; the fourth column shows the hypergeometric P-value of observing similar numbers by chance. The two modes segregate the binding sites into two motifs (left bottom), which together capture these dependencies.
