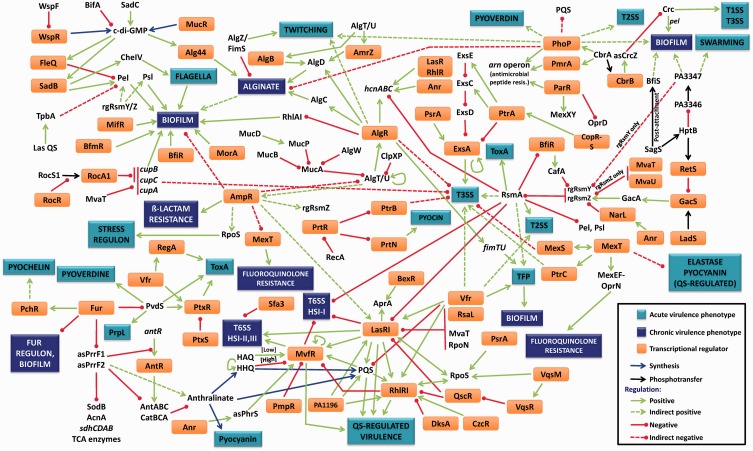
Figure 1.
The P. aeruginosa virulence regulatory network. The pathogenic potential of P. aeruginosa is dictated by multiple virulence systems that are regulated transcriptionally, post-transcriptionally and post-translationally. The central mechanism for P. aeruginosa virulence regulation is QS, which controls expression of many virulence factors in a population density-dependent manner. Key activators of this system are LasR, RhlR, MvfR, VqsR, the cAMP receptor protein Vfr and the stationary phase σ factor RpoS. Las system repressors include RsaL, the H-NS protein MvaT, the σ factor RpoN and the sRNA-binding protein RsmA, whereas others like QscR repress both the Las and Rhl systems. Other regulators such as AmpR affect QS genes by an unknown mechanism. QS plays a role in regulating critical pathogenic mechanisms, including biofilm formation, secretion systems, production of numerous virulence factors, efflux pumps, antibiotic resistance and motility. Acute P. aeruginosa infections can lead to chronic infections in response to largely unidentified signals. A key regulatory pathway that controls this lifestyle switch is the RetS–LadS–GacSA–RsmA pathway. RetS and LadS are hybrid sensor proteins that, in response to external signals, either activate or repress the GacSA TCS. The GacA regulator then activates transcription of two rgRNAs, rgRsmZ and rgRsmY that sequester and inhibit activity of the sRNA-binding protein, RsmA. RsmA is a key activator/repressor that post-transcriptionally regulates numerous acute and chronic infection phenotypes, including multiple QS-regulated virulence factors, biofilm formation, Type 2, Type 3 and Type 6 secretion systems and motility. Another major phenotypic change associated with the switch from acute to chronic phases of infection is the formation of biofilm. This is associated with extensive changes in transcription. Three key TCS involved in activating biofilm formation are BfiSR, MifR and BfmSR. Cyclic-di-GMP is another major player influencing this process, whose levels are controlled by diguanylate cyclases and phosphodiesterases. QS and the cup genes enhance biofilms, whereas regulators like AmpR repress it. An important component of P. aeruginosa biofilms are extracellular polysaccharides, such as alginate, Pel and Psl. Alginate production is under the control of the master regulator ECF AlgT/U, whose activity is regulated transcriptionally by AmpR, post-translationally by MucA and MucB and by regulated intermembrane proteolysis involving MucP, AlgW, ClpXP and others. AlgT/U activates the alginate biosynthetic operon through AlgR, AlgB and AmrZ. In addition, biofilm formation is also affected by iron concentration, a process governed by the master repressor of iron uptake, Fur. Fur controls uptake of iron by regulating the σ factor PvdS, thereby modulating sidephore levels. Fur also modulates transcription of two key regulatory RNAs, asPrrF1 and asPrrF2. These two sRNAs are involved not only in regulating iron-uptake-related genes but also enzymes of the trichloroacetic acid cycle and genes involved in anthranilate synthesis. Anthralinate, a precursor for synthesis of PQS, is a key regulatory molecule of the PQS signalling system in P. aeruginosa, which is involved in expression of QS-regulated virulence factors. Details on the individual interactions and the appropriate references can be found in the text. Some of the interactions labelled as indirect are regulated by unknown mechanisms and warrant further investigation. In the figure, some regulators and phenotypes have been mentioned more than once.