Figure 1.
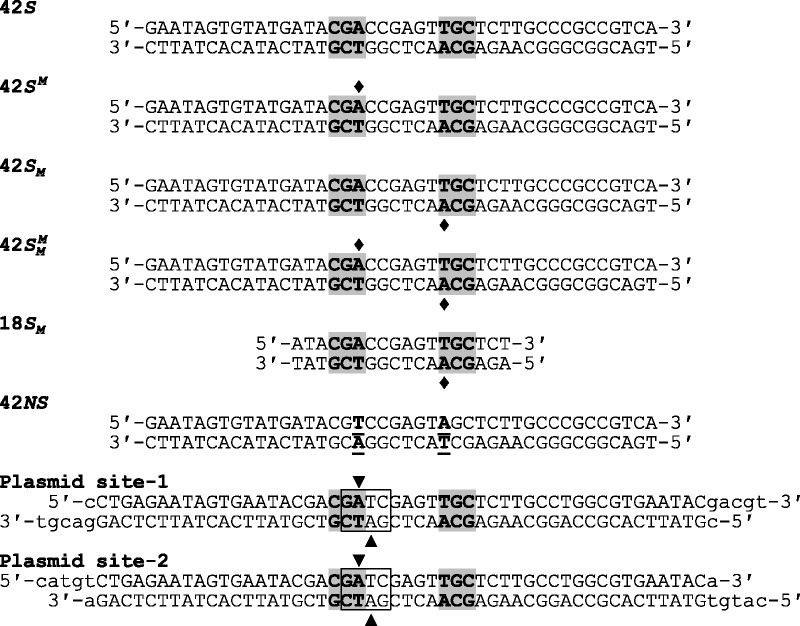
Substrates. The oligoduplexes used in this study are named on the left and have the sequences indicated. In all cases except for 42NS, the recognition sequence for BcgI is shaded in grey and the diamonds mark the locations where the duplex in question carried m6A in place of adenine. In 42NS, the two adenine residues modified by the BcgI MTase were both changed to thymines (the underlined bp, in bold) to leave a non-specific sequence for BcgI. In the oligoduplexes used in the construction of plasmids pRMS01 and pRMS02, the box encloses a Dam site (GATC) that overlaps the left-hand half of the recognition sequence for BcgI. In a dam+ strain, the two adenines within the Dam site, noted by the inverted and upright triangles in top and bottom strands, respectively, are both converted to m6A. The inverted triangle also specifies the adenine that is shared by the BcgI site so that, when purified from a dam+ strain, this overlapping arrangement results in HM DNA at the BcgI site. Dam methylation of the bottom strand, at the adenine marked by the upright triangle, occurs within the 6 bp spacer of random sequence in the BcgI site, distinct from the site of bottom-strand methylation by BcgI.