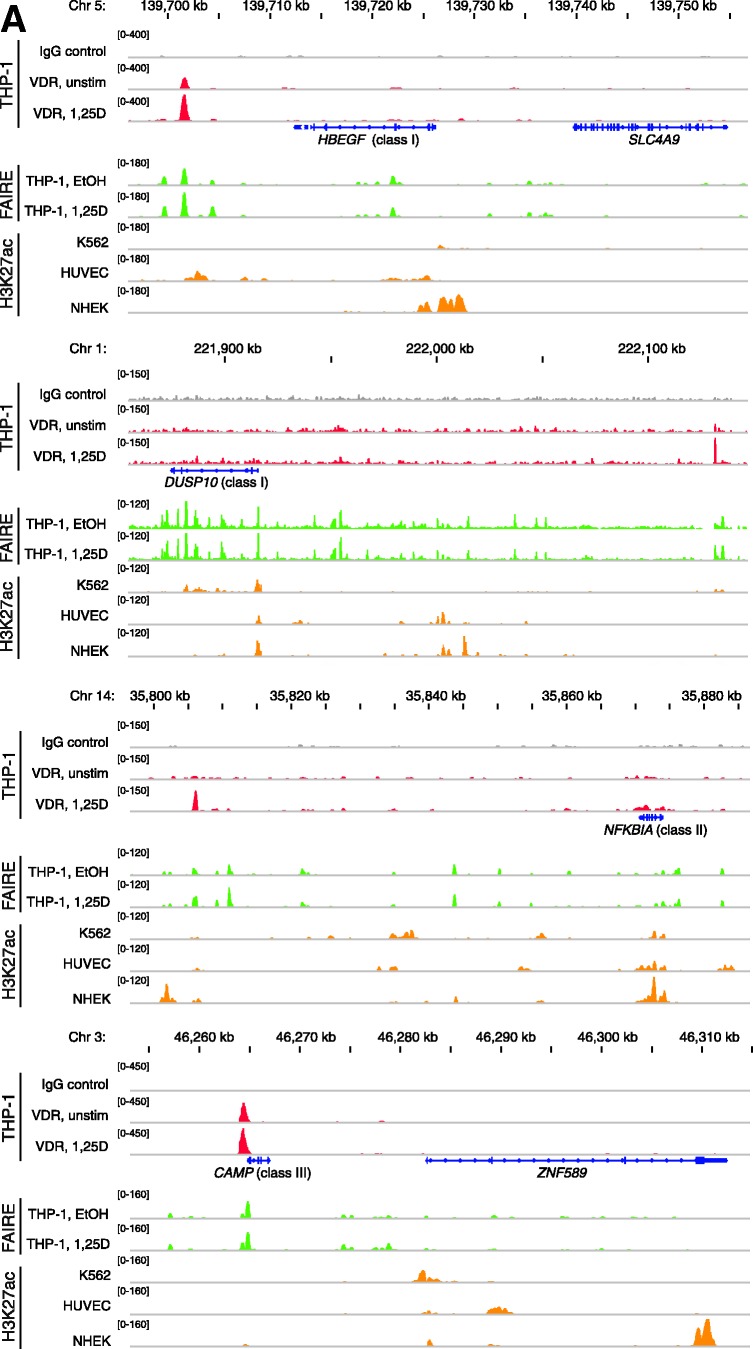
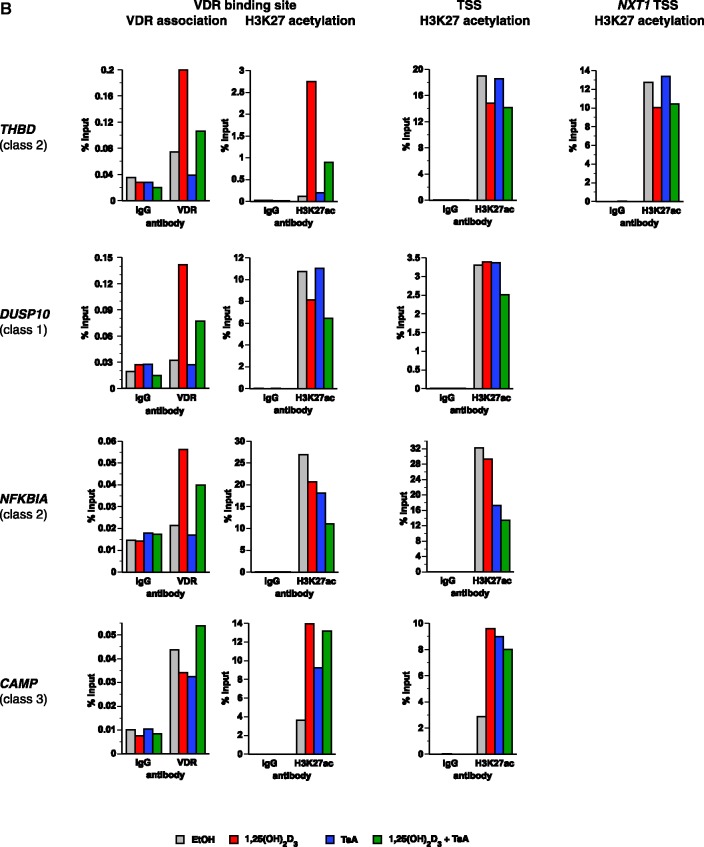
Figure 3.
VDR association, histone acetylation and sites of open chromatin for selected loci. (A) Loci of the genes HBEGF, DUSP10, NFKBIA and CAMP. The peak tracks for the four different gene loci show data from a VDR ChIP-seq experiment in THP-1 cells (34) (IgG control in grey, VDR binding in unstimulated cells and VDR binding after 40 min 1α,25(OH)2D3 (1,25D) treatment in red), FAIRE-seq data from THP-1 cells (EtOH- and 1α,25(OH)2D3-treated for 100 min, green), and H3K27ac ChIP-seq data from the ENCODE (45) cell lines K562, HUVEC and NHEK (orange) as a sign of histone acetylation. The gene structures are shown in blue. (B) VDR association, H3K27 acetylation and unspecific IgG binding at VDR binding sites and TSS regions of four VDR target genes, determined by ChIP-qPCR after 40 min treatments of THP-1 cells with solvent, 1α,25(OH)2D3, TsA or the combination of both. Each graph is a representative of four independent experiments. Please note that the best VDR binding sites of the THBD and NFKBIA loci chosen for this assay are those close to the NXT1 and PSMA6 genes, respectively (Figures 1A and 3A).