Table 1.
Parameter values used to simulate the restrained bead-chain polymer model of chromatin and to provide a physically realistic approximation of the mechanical properties of chromatin, as currently known from experiments
| Parameter | Symbol | Reduced units | SI units |
|---|---|---|---|
| Thermal energya | 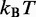 |
1.0 | 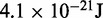 |
| Bead massb | m | 1.0 | 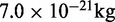 |
| Lennard–Jones size parameter |  |
1.0 | 30 nm |
| Lennard–Jones energy parameter | 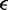 |
 |
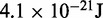 |
| Bead separation |  |
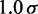 |
30 nm |
| Contact distancec | 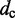 |
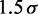 |
45 nm |
| Bond spring constantd | 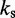 |
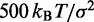 |
 |
| Persistence lengthe | 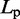 |
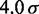 |
120 nm |
| Bending energy constant | 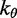 |
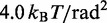 |
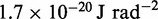 |
| Time step/damping constantf | 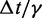 |
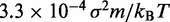 |
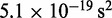 |
aEnergy per bead per degree of freedom at T = 300 K.
bRepresentative value based on the experimental measurement of 23.3 MDa for a 15.5-kb fragment of 30-nm chromatin upstream of the chicken 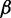 -globin locus (42).
-globin locus (42).
cFollowing Rosa et al. (35), equivalent to assuming that contacts between chromatin fibers are mediated by proteins of 15-nm diameter.
dFrom experiments, the stretching modulus is 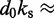 5–150 pN (43), hence
5–150 pN (43), hence 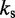 ranges from
ranges from 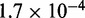 to
to 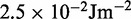 .
.
eFrom experiments,  30 – 200 nm (43).
30 – 200 nm (43).
fTo maximize conformation sampling efficiency, we used the largest value of 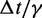 found to maintain stability of the BD simulations. A lower bound for
found to maintain stability of the BD simulations. A lower bound for  can be estimated by considering a chromatin sphere of radius r = 15 nm and using
can be estimated by considering a chromatin sphere of radius r = 15 nm and using 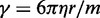 with the viscosity of water
with the viscosity of water 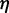 = 890 µPa s at 25°C and 1 bar (44). Then,
= 890 µPa s at 25°C and 1 bar (44). Then, 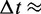 18 ns.
18 ns.
