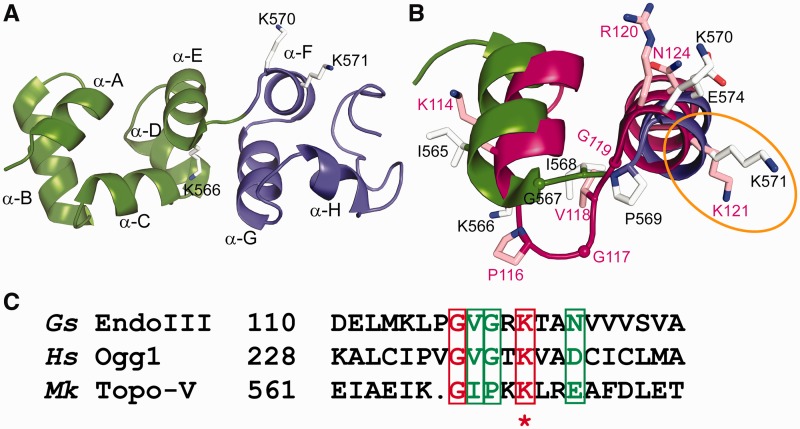
Figure 4.
Superposition of the Topo-78 structure onto EndoIII [PDB ID: 1ORN (29)]. (A) Close-up view of (HhH)2 domains 5 (green) and 6 (blue), forming the AP lyase active site. The successive helices are named from A to J. K566 is the last residue of helix α-E, and K570 and K571 are the first two residues of helix α-F. (B) Comparison of the DNA repair active sites of Topo-78 (green and blue) and EndoIII (magenta). Residues K566, K570 and K571 of Topo-78 match L115, R120 and K121 of EndoIII respectively, with K114 immediately adjacent to K566. The location of these residues in the helices is very similar in both enzymes despite the longer loop connecting the two helices in EndoIII. (C) Sequence comparison of the HhH motifs from various proteins involved in DNA repair. The region for comparison was taken from Doherty et al. (30). Gs EndoIII, G. stearothermophilus EndoIII; Hs Ogg1, human 8-oxoguanine glycosylase; Mk Topo-V, M. kandleri Topo-V. Sequence analysis shows that the equivalent lysine of Topo-78, K571 (marked with an asterisk), is the nucleophile in all the proteins compared.