Figure 1.
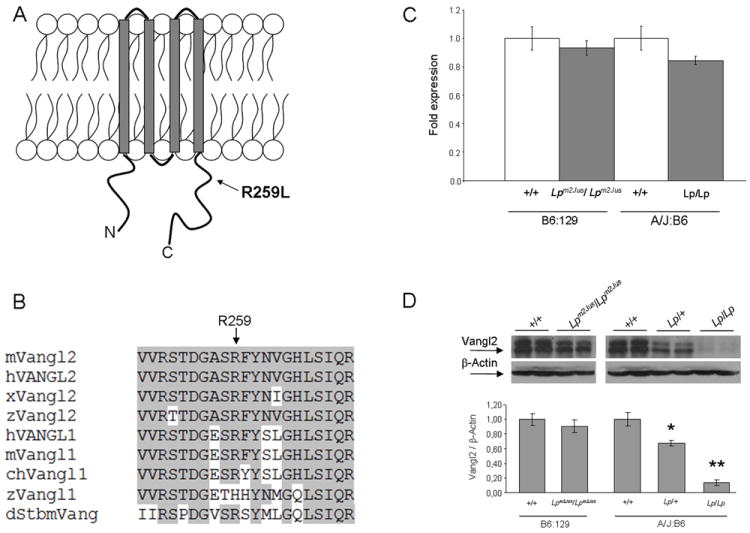
Molecular characterization of the Lpm2Jus mutant. Panel A. Diagram of the Vangl2 protein structure with the approximate position of the R259L mutation identified in Lpm2Jus indicated by an arrow. Panel B. Partial alignment of mouse Vangl2 with 8 other Vangl/Stbm sequences. Residues conserved between Vangl2 and other family members are highlighted. The R259L variant affects a highly conserved amino acid residue (indicated by an arrow). Accession numbers: mouse Vangl2 (mVangl2), NP_277044; human VANGL2 (hVANGL2), NP_065068; frog Vangl2 (xVangl2), AAK70879; zebrafishVangl2 (zVangl2), NP_705960; human VANGL1 (hVANGL1), NP_620409; mouse Vangl1 (mVangl1), NP_808213; chicken Vangl1 (chVangl1), XP_001232441; zebrafishVangl1 (zVangl1), AAQ84560; and Drosophila Stbm (dStbm), NP_477177. Panel C. Real- time PCR showing the relative levels of Vangl2 mRNA compared with Hprt transcripts in brain tissues of E16.5 wild-type and mutant embryos. Panel D. Western analysis of Vangl2 protein levels in brain tissues of E16.5 wild-type and mutant embryos. β-actin was used as an internal control. Top, western blots of two representative embryos per group are shown. Bottom, a histogram showing the relative levels of Vangl2 protein compared with β-actin. A significant difference was detected between wild-type and Lp/+ (*P<0.001) and between wild-type and Lp/Lp embryos (**P<0.0001).
