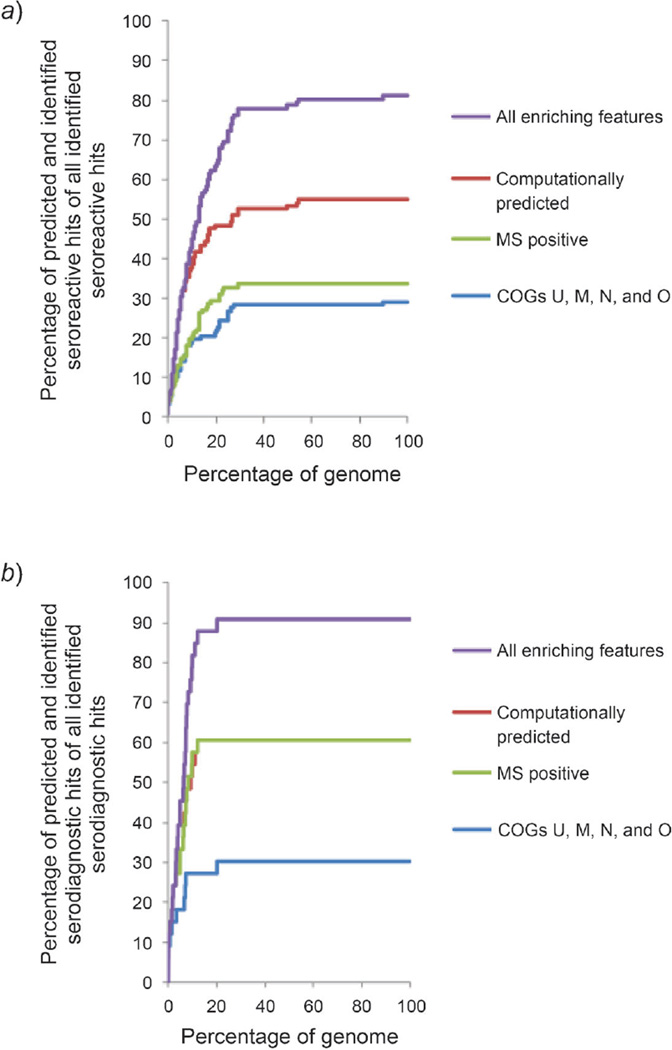
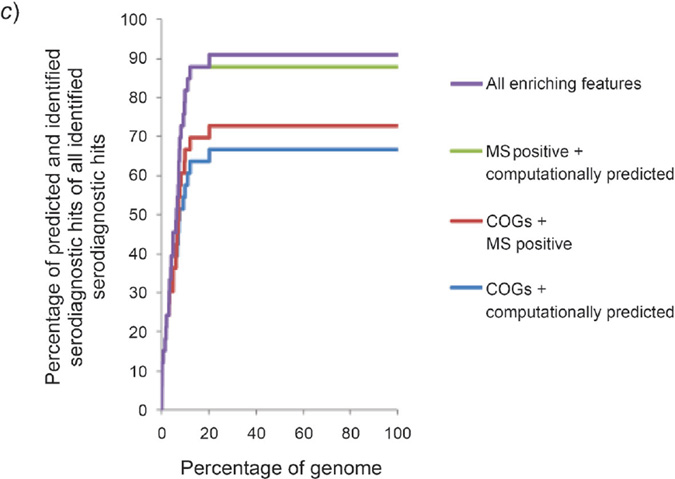
Fig. 2. Categorized enriching features on prediction of seroreactive or serodiagnostic antigens based on naïve Bayes ranking.
a) Prediction of categorized enriching features on seroreactive antigens. Within the top 30% of the ranked genome, COGs (COG U, M, N, and O), computationally predicted features, the MS positive feature, and the ten enriching features altogether could predict 30, 52, 34, and 78% of all seroreactive hits, respectively. b) Prediction of categorized enriching features on serodiagnostic antigens. Within the top 20% of the ranked genome, COGs (COG U, M, N, and O), computationally predicted features, the MS positive feature, and the ten enriching features altogether could predict 30, 61, 61, and 91% of all serodiagnostic hits, respectively. c) Prediction of paired categorized enriching features on serodiagnostic antigens. Pairing two of the categories of the enriching features significantly enhanced the prediction on serodiagnostic antigens compared to prediction from an individual category of enriching features.