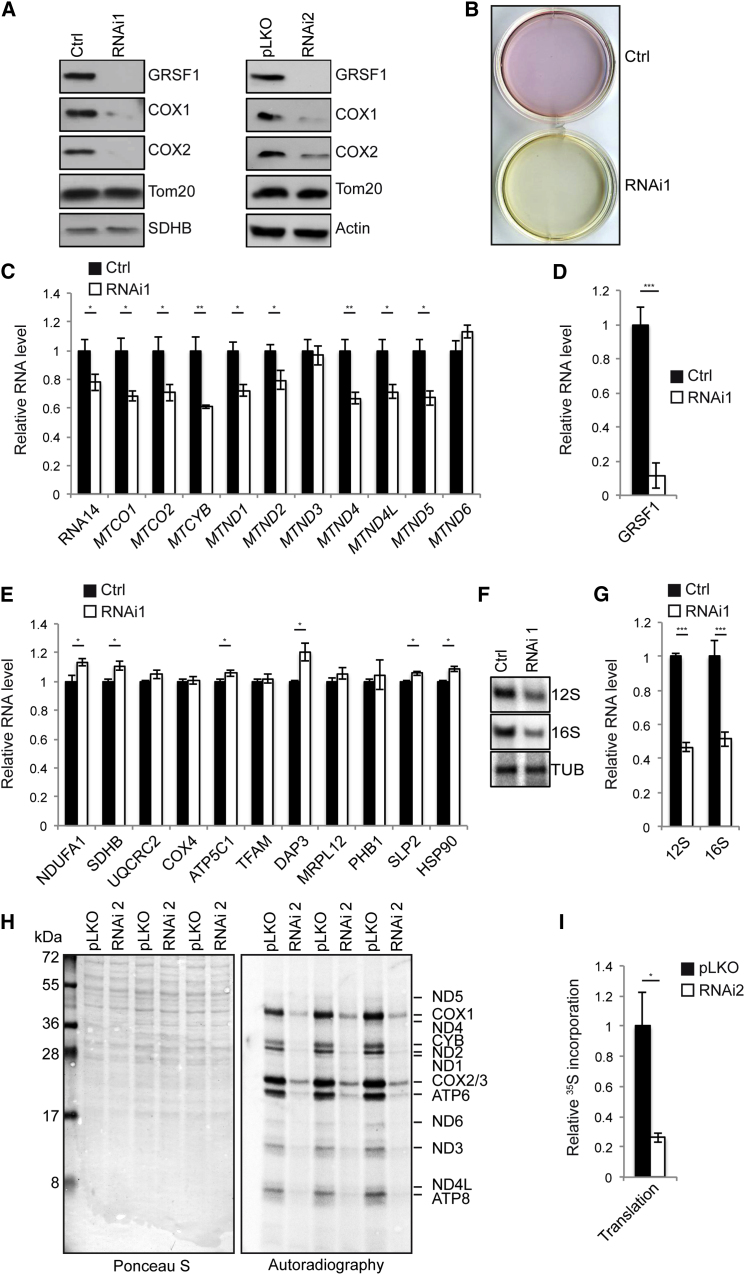
Figure 3.
GRSF1 Is Required for Mitochondrial Gene Expression
(A) Left panel, immunoblot analysis of 143B cells treated with control (Ctrl) or GRSF1 RNAi1. Right panel, immunoblot analysis of 143B cells infected with lentiviruses carrying an empty vector (pLKO) or a shRNA (RNAi2) against GRSF1. COX1 and COX2 are mitochondrially encoded proteins. SDHB and Tom20 are nuclear-encoded mitochondrial proteins. Actin is a nuclear-encoded cytosolic protein.
(B) Acidification of the culture medium in GRSF1 RNAi1-treated 143B cells. The number of cells was the same in both conditions. Similar results were obtained in GRSF1 RNAi2-infected cells (data not shown).
(C) NanoString analysis of 11 mitochondrially encoded ORFs in control (Ctrl) or GRSF1 RNAi1-treated 143B cells. RNA14 corresponds to bicistronic MTATP8-MTATP6 RNA. Data are shown as mean ± SEM (n = 3).
(D) NanoString analysis of GRSF1 in control (Ctrl) or GRSF1 RNAi1-treated 143B cells. Data are shown as mean ± SEM (n = 3).
(E) NanoString analysis of 11 nuclear-encoded proteins in control (Ctrl) or GRSF1 RNAi1-treated 143B cells. Data are shown as mean ± SEM (n = 3).
(F) Northern blot analysis of 12S and 16S rRNA in control (Ctrl) or GRSF1 RNAi1-treated cells. TUB, tubulin.
(G) Phosphorimager quantification of (E). Data are shown as mean ± SEM (n = 3).
(H) 35S-labeling of mitochondrial translation of 143B cells infected with lentiviruses carrying an empty vector (pLKO) or a shRNA (RNAi2) against GRSF1.
(I) Phosphorimager quantification of (H). Data are normalized to the Ponceau S intensity and are shown as mean ± SEM (n = 4).