Figure 7.
The Kinetics of RNA Release from MRGs Is Dependent on GRSF1
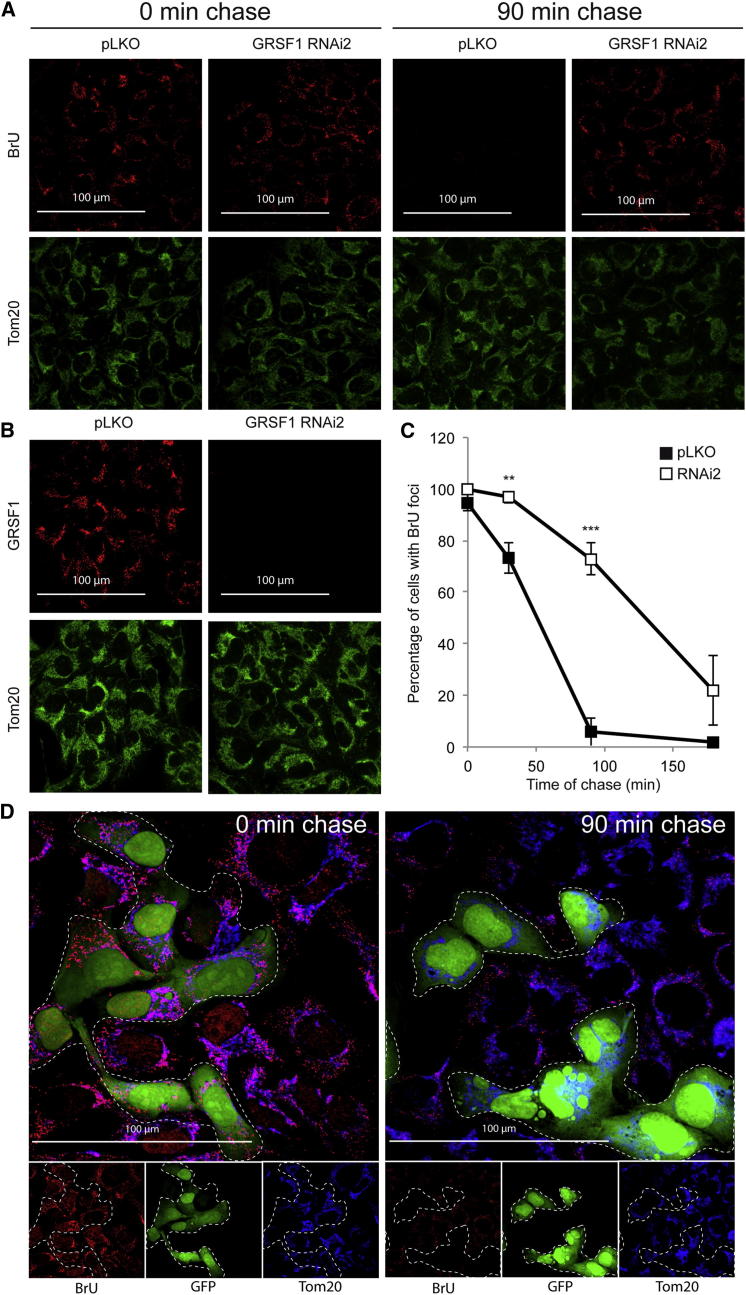
(A) Confocal analysis of 143B infected with pLKO or GRSF1 RNAi2-carrying lentiviruses after 60 min of BrU pulse and 0 or 90 min of chase with uridine. The parameters used for the entire analysis were identical for both conditions. Note that the thresholds were set up so that only the BrU foci could be visualized.
(B) Confocal analysis of endogenous GRSF1 and Tom20 in pLKO or GRSF1 RNAi2-infected cells. The parameters used for the entire analysis were identical for both conditions.
(C) Number of cells with BrU foci after 60 min of BrU pulse and up to 180 min of chase with uridine in 143B cells infected with pLKO or GRSF1 RNAi2-carrying lentiviruses. Cells were counted using widefield microscopy. Data are shown as mean ± SEM (n = 3).
(D) Confocal analysis of BrU pulse chase of mixed control (green) and GRSF1 RNAi2 cells. Control 143B cells were infected with lentiviruses carrying an empty pLKO vector and lentiviruses carrying the GFP gene (pLKO + GFP), whereas GRSF1-depleted cells were infected with lentiviruses expressing GRSF1 RNAi2. All control cells were found to express GFP (data not shown). The two cell populations were mixed, selected with puromycin for 24 hr to eliminate those cells that would not have been infected by lentiviruses, and incubated with BrU for 60 min before chase with uridine. Cells were immunostained with anti-BrU and anti-Tom20 antibodies.