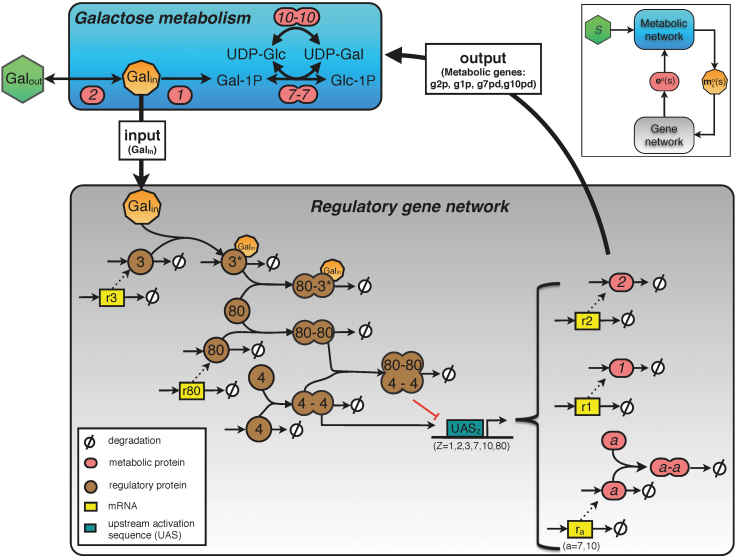
Figure 2. Modular representation of the galactose network and it's regulatory interactions in yeast.
Shown are the inputs and outputs of the galactose metabolism and galactose regulatory network, using a similar representation as in Figure 1A. Galactose metabolism (shown in blue) consists of four metabolic enzymes (gal2p, gal1p, gal7pd, gal10pd, shown in red). External galactose (Galout, green), is imported by gal2p, resulting in intracellular galactose (Galin, orange), which is further metabolised into glucose-1-phosphate (Glc-1P) by the enzymes gal7pd and gal10pd. Galin is needed for activation of the galactose regulatory network by binding to gal3p. Within this network, a distinction can be made between the regulatory proteins, gal3p, gal80p, gal4p (brown) and structural proteins (metabolic enzymes; red). Transcription of all genes is dependent on the concentration of gal4p dimer (gal4pd) and the number of gal4dp binding sites that the upstream activating sequences (UAS's) possess. The resulting mRNA's are shown in yellow. Degradation of every mRNA and protein is the net effect of intrinsic degradation and the growth rate dependent dilution.