Abstract
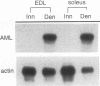
Although most skeletal muscle genes are expressed at similar levels in electrically active, innervated muscle and in electrically inactive, denervated muscle, a small number of genes, including those encoding the acetylcholine receptor, N-CAM, and myogenin, are expressed at significantly higher levels in denervated than in innervated muscle. The mechanisms that mediate electrical activity-dependent gene regulation are not understood, but these mechanisms are likely to be responsible, at least in part, for the changes in muscle structure and function that accompany a decrease in myofiber electrical activity. To understand how muscle activity regulates muscle structure and function, we used a subtractive-hybridization and cloning strategy to identify and isolate genes that are expressed preferentially in innervated or denervated muscle. One of the genes which we found to be regulated by electrical activity is the recently discovered acute myeloid leukemia 1 (AML1) gene. Disruption and translocation of the human AML1 gene are responsible for a form of acute myeloid leukemia. AML1 is a DNA-binding protein, but its normal function is not known and its expression and regulation in skeletal muscle were not previously appreciated. Because of its potential role as a transcriptional mediator of electrical activity, we characterized expression of the AML1 gene in innervated, denervated, and developing skeletal muscle. We show that AML1 is expressed at low levels in innervated skeletal muscle and at 50- to 100-fold-higher levels in denervated muscle. Four AML1 transcripts are expressed in denervated muscle, and the abundance of each transcript increases after denervation. We transfected C2 muscle cells with an expression vector encoding AML1, tagged with an epitope from hemagglutinin, and we show that AML1 is a nuclear protein in muscle. AML1 dimerizes with core-binding factor beta (CBF beta), and we show that CGF beta is expressed at high levels in both innervated and denervated skeletal muscle. PEBP2 alpha, which is structurally related to AML1 and which also dimerizes with CBF beta, is expressed at low levels in skeletal muscle and is up-regulated only weakly by denervation. These results are consistent with the idea that AML1 may have a role in regulating gene expression in skeletal muscle.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson M. J., Cohen M. W. Nerve-induced and spontaneous redistribution of acetylcholine receptors on cultured muscle cells. J Physiol. 1977 Jul;268(3):757–773. doi: 10.1113/jphysiol.1977.sp011880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atomi Y., Yamada S., Nishida T. Early changes of alpha B-crystallin mRNA in rat skeletal muscle to mechanical tension and denervation. Biochem Biophys Res Commun. 1991 Dec 31;181(3):1323–1330. doi: 10.1016/0006-291x(91)92083-v. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. C., Ogawa E., Maruyama M., Oka H., Satake M., Shigesada K., Jenkins N. A., Gilbert D. J., Copeland N. G., Ito Y. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994 May;14(5):3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. C., Yamaguchi-Iwai Y., Ogawa E., Maruyama M., Inuzuka M., Kagoshima H., Shigesada K., Satake M., Ito Y. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993 Mar;8(3):809–814. [PubMed] [Google Scholar]
- Block N. E., Menick D. R., Robinson K. A., Buse M. G. Effect of denervation on the expression of two glucose transporter isoforms in rat hindlimb muscle. J Clin Invest. 1991 Nov;88(5):1546–1552. doi: 10.1172/JCI115465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes J. P. Assays for cholinergic properties in cultured rat Schwann cells. Proc R Soc Lond B Biol Sci. 1984 Jul 23;222(1226):121–134. doi: 10.1098/rspb.1984.0053. [DOI] [PubMed] [Google Scholar]
- Burden S. J. Synapse-specific gene expression. Trends Genet. 1993 Jan;9(1):12–16. doi: 10.1016/0168-9525(93)90066-Q. [DOI] [PubMed] [Google Scholar]
- Castelló A., Cadefau J., Cussó R., Testar X., Hesketh J. E., Palacín M., Zorzano A. GLUT-4 and GLUT-1 glucose transporter expression is differentially regulated by contractile activity in skeletal muscle. J Biol Chem. 1993 Jul 15;268(20):14998–15003. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Covault J., Merlie J. P., Goridis C., Sanes J. R. Molecular forms of N-CAM and its RNA in developing and denervated skeletal muscle. J Cell Biol. 1986 Mar;102(3):731–739. doi: 10.1083/jcb.102.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A., Tighe J. E., Calabi F. Leukaemia/Drosophila homology. Nature. 1992 Apr 9;356(6369):484–484. doi: 10.1038/356484b0. [DOI] [PubMed] [Google Scholar]
- Donoghue M. J., Alvarez J. D., Merlie J. P., Sanes J. R. Fiber type- and position-dependent expression of a myosin light chain-CAT transgene detected with a novel histochemical stain for CAT. J Cell Biol. 1991 Oct;115(2):423–434. doi: 10.1083/jcb.115.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclert A., Piette J., Changeux J. P. Influence of innervation of myogenic factors and acetylcholine receptor alpha-subunit mRNAs. Neuroreport. 1991 Jan;2(1):25–28. doi: 10.1097/00001756-199101000-00006. [DOI] [PubMed] [Google Scholar]
- Eftimie R., Brenner H. R., Buonanno A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1349–1353. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P., Gao J., Chang K. S., Look T., Whisenant E., Raimondi S., Lasher R., Trujillo J., Rowley J., Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992 Oct 1;80(7):1825–1831. [PubMed] [Google Scholar]
- Evans S., Goldman D., Heinemann S., Patrick J. Muscle acetylcholine receptor biosynthesis. Regulation by transcript availability. J Biol Chem. 1987 Apr 5;262(10):4911–4916. [PMC free article] [PubMed] [Google Scholar]
- Gatchalian C. L., Schachner M., Sanes J. R. Fibroblasts that proliferate near denervated synaptic sites in skeletal muscle synthesize the adhesive molecules tenascin(J1), N-CAM, fibronectin, and a heparan sulfate proteoglycan. J Cell Biol. 1989 May;108(5):1873–1890. doi: 10.1083/jcb.108.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. G., Covault J. Neural regulation of N-cadherin gene expression in developing and adult skeletal muscle. J Neurosci. 1992 Dec;12(12):4677–4687. doi: 10.1523/JNEUROSCI.12-12-04677.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Z. W., Sanes J. R. Synaptic structure and development: the neuromuscular junction. Cell. 1993 Jan;72 (Suppl):99–121. doi: 10.1016/s0092-8674(05)80031-5. [DOI] [PubMed] [Google Scholar]
- Hallberg B., Schmidt J., Luz A., Pedersen F. S., Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991 Aug;65(8):4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgren M. E., Squinto S. P., Davis H. L., Parry D. J., Boulton T. G., Heck C. S., Zhu Y., Yancopoulos G. D., Lindsay R. M., DiStefano P. S. Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell. 1994 Feb 11;76(3):493–504. doi: 10.1016/0092-8674(94)90113-9. [DOI] [PubMed] [Google Scholar]
- Hughes S. M., Taylor J. M., Tapscott S. J., Gurley C. M., Carter W. J., Peterson C. A. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993 Aug;118(4):1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Jennings C. G., Burden S. J. Development of the neuromuscular synapse. Curr Opin Neurobiol. 1993 Feb;3(1):75–81. doi: 10.1016/0959-4388(93)90038-z. [DOI] [PubMed] [Google Scholar]
- Kamachi Y., Ogawa E., Asano M., Ishida S., Murakami Y., Satake M., Ito Y., Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990 Oct;64(10):4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania M. A., Bonner A. S., Duffy J. B., Gergen J. P. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990 Oct;4(10):1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- Labeit S., Barlow D. P., Gautel M., Gibson T., Holt J., Hsieh C. L., Francke U., Leonard K., Wardale J., Whiting A. A regular pattern of two types of 100-residue motif in the sequence of titin. Nature. 1990 May 17;345(6272):273–276. doi: 10.1038/345273a0. [DOI] [PubMed] [Google Scholar]
- Laufer R., Changeux J. P. Activity-dependent regulation of gene expression in muscle and neuronal cells. Mol Neurobiol. 1989 Spring-Summer;3(1-2):1–53. doi: 10.1007/BF02935587. [DOI] [PubMed] [Google Scholar]
- Liu P., Tarlé S. A., Hajra A., Claxton D. F., Marlton P., Freedman M., Siciliano M. J., Collins F. S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993 Aug 20;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- McMahan U. J. The agrin hypothesis. Cold Spring Harb Symp Quant Biol. 1990;55:407–418. doi: 10.1101/sqb.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- Melnikova I. N., Crute B. E., Wang S., Speck N. A. Sequence specificity of the core-binding factor. J Virol. 1993 Apr;67(4):2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie J. P., Isenberg K. E., Russell S. D., Sanes J. R. Denervation supersensitivity in skeletal muscle: analysis with a cloned cDNA probe. J Cell Biol. 1984 Jul;99(1 Pt 1):332–335. doi: 10.1083/jcb.99.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie J. P., Sanes J. R. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibres. Nature. 1985 Sep 5;317(6032):66–68. doi: 10.1038/317066a0. [DOI] [PubMed] [Google Scholar]
- Meyers S., Downing J. R., Hiebert S. W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993 Oct;13(10):6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Kozu T., Shimizu K., Enomoto K., Maseki N., Kaneko Y., Kamada N., Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993 Jul;12(7):2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastuk M. A., Fallon J. R. Agrin and the molecular choreography of synapse formation. Trends Neurosci. 1993 Feb;16(2):72–76. doi: 10.1016/0166-2236(93)90020-m. [DOI] [PubMed] [Google Scholar]
- Neville C. M., Schmidt M., Schmidt J. Response of myogenic determination factors to cessation and resumption of electrical activity in skeletal muscle: a possible role for myogenin in denervation supersensitivity. Cell Mol Neurobiol. 1992 Dec;12(6):511–527. doi: 10.1007/BF00711232. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka M., Maruyama M., Satake M., Naito-Fujimoto M., Ito Y., Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993 May;194(1):314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Maruyama M., Kagoshima H., Inuzuka M., Lu J., Satake M., Shigesada K., Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D., Staron R. S. Cellular and molecular diversities of mammalian skeletal muscle fibers. Rev Physiol Biochem Pharmacol. 1990;116:1–76. doi: 10.1007/3540528806_3. [DOI] [PubMed] [Google Scholar]
- Reynolds M. L., Woolf C. J. Terminal Schwann cells elaborate extensive processes following denervation of the motor endplate. J Neurocytol. 1992 Jan;21(1):50–66. doi: 10.1007/BF01206897. [DOI] [PubMed] [Google Scholar]
- Roebroek A. J., van de Velde H. J., Van Bokhoven A., Broers J. L., Ramaekers F. C., Van de Ven W. J. Cloning and expression of alternative transcripts of a novel neuroendocrine-specific gene and identification of its 135-kDa translational product. J Biol Chem. 1993 Jun 25;268(18):13439–13447. [PubMed] [Google Scholar]
- Schiaffino S., Gorza L., Sartore S., Saggin L., Ausoni S., Vianello M., Gundersen K., Lømo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989 Jun;10(3):197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Selden R. F., Howie K. B., Rowe M. E., Goodman H. M., Moore D. D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol Cell Biol. 1986 Sep;6(9):3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A. M., Burden S. J. An E box mediates activation and repression of the acetylcholine receptor delta-subunit gene during myogenesis. Mol Cell Biol. 1993 Sep;13(9):5133–5140. doi: 10.1128/mcb.13.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A. M., Hoppe P., Burden S. J. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992 Mar;114(3):545–553. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Tang J., Jo S. A., Burden S. J. Separate pathways for synapse-specific and electrical activity-dependent gene expression in skeletal muscle. Development. 1994 Jul;120(7):1799–1804. doi: 10.1242/dev.120.7.1799. [DOI] [PubMed] [Google Scholar]
- Voytik S. L., Przyborski M., Badylak S. F., Konieczny S. F. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev Dyn. 1993 Nov;198(3):214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- Wang S. W., Speck N. A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992 Jan;12(1):89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang Q., Crute B. E., Melnikova I. N., Keller S. R., Speck N. A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993 Jun;13(6):3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Brown D. D. A gene expression screen. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D. F., Hughes S. R. Developmentally regulated cDNA expressed exclusively in neural tissue. Brain Res Mol Brain Res. 1991 Apr;10(1):33–41. doi: 10.1016/0169-328x(91)90053-z. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Witzemann V., Sakmann B. Differential regulation of MyoD and myogenin mRNA levels by nerve induced muscle activity. FEBS Lett. 1991 May 6;282(2):259–264. doi: 10.1016/0014-5793(91)80490-t. [DOI] [PubMed] [Google Scholar]
- Yang J. S., Sladky J. T., Kallen R. G., Barchi R. L. TTX-sensitive and TTX-insensitive sodium channel mRNA transcripts are independently regulated in adult skeletal muscle after denervation. Neuron. 1991 Sep;7(3):421–427. doi: 10.1016/0896-6273(91)90294-a. [DOI] [PubMed] [Google Scholar]