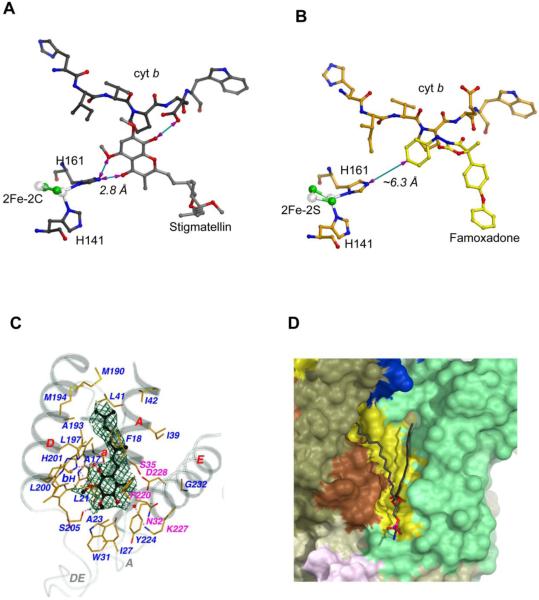
Figure 3. Binding of substrate, inhibitor and lipid molecules to the cyt bc1 complex.
(A) Hydrogen bonding interaction between stigmatellin and the ISP. In this figure, much of the protein structures of cyt b and ISP are omitted for clarity to illustrate the H-bond between stigmatellin and the ISC. All residues, the ISC, and stigmatellin are shown as ball-and-stick models and as labeled. The distance between BTH161, one of the 2Fe-2S ligand, and stigmatellin is shown as 2.8 Å. (B) No hydrogen bonding is observed between famoxadone and the ISP. Binding of famoxadone arrests the ISP-ED motion. But the closest distance between BTH161 and the ISP is >6 Å. (C) Interaction of the protein environment at the QN site of the cyt b subunit with bound substrate ubiquinone with two isoprenoid repeats. Secondary structure elements surrounding the QN pocket, including portions of the N-terminal helix a, TM helices A, D, and E, and extra-membrane loops A and DE, are shown and are labeled. Residues interacting with bound substrate and the bH heme are drawn in stick models and are labeled with carbon atoms in yellow, nitrogen in blue, oxygen in red, and iron in orange. H-bonds are indicated with pinkish dotted lines. Water molecules are shown as isolated red balls. The residues that are with magenta labels confer inhibitor resistance. The substrate ubiquinone, caged in Fo-Fc electron densities calculated with refined phases after ligand being omitted and contoured at the 3σ level in dark green, are drawn as ball-and-stick models with carbon atoms in black, nitrogen in light blue, and oxygen in red. Additionally, the two bound water molecules are enclosed in the Fo-Fc electron density in cyan calculated with refined phases obtained with the waters omitted and contoured at 3σ. (D) Binding environment of the bound lauryl oleoyl phosphatidyl ethanolamine (PE) in the structure of Rsbc1. The modeled lipid is located near the N-side and its binding environment is shown.