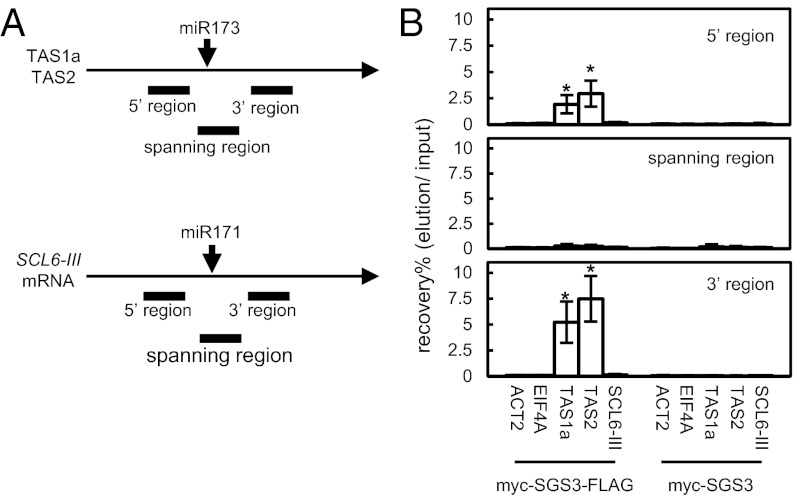
Fig. 1.
Complex containing SGS3 and miR173-cleaved 5′ and 3′ fragments is formed during tasiRNA production. (A) Diagrams of TAS1a, TAS2, and SCL6-III transcripts. The 5′ regions, spanning regions, and 3′ regions correspond to upstream, spanning, and downstream of each miRNA cleavage site, respectively. (B) Analysis of SGS3-bound RNAs. Floral tissue extracts were prepared from transgenic plants from a self-pollinated primary transgenic plant expressing myc- and FLAG-double-tagged SGS3 (myc-SGS3-FLAG) or myc-tagged SGS3 (myc-SGS3) and subjected to stepwise glycerol gradient centrifugation. FLAG-tagged SGS3 was immunoprecipitated with anti-FLAG antibody from the TAS2 RNA-rich fractions (Fig. S1, fraction 8). RNA was purified from the fractions, and the amounts of the 5′ regions (Top), spanning regions (Middle), and 3′ regions (Bottom) of TAS1a, TAS2, and SCL6-III RNA were quantified by RT-PCR. Transcripts of ACT2 and EIF4A were used as controls. SDs were calculated from three biological repeats. Asterisks indicate that values are significantly different from those of tag minus controls (*P < 0.05 by Tukey–Kramer test). Similar results were obtained in another experiment using an independent transgenic line for each tag.