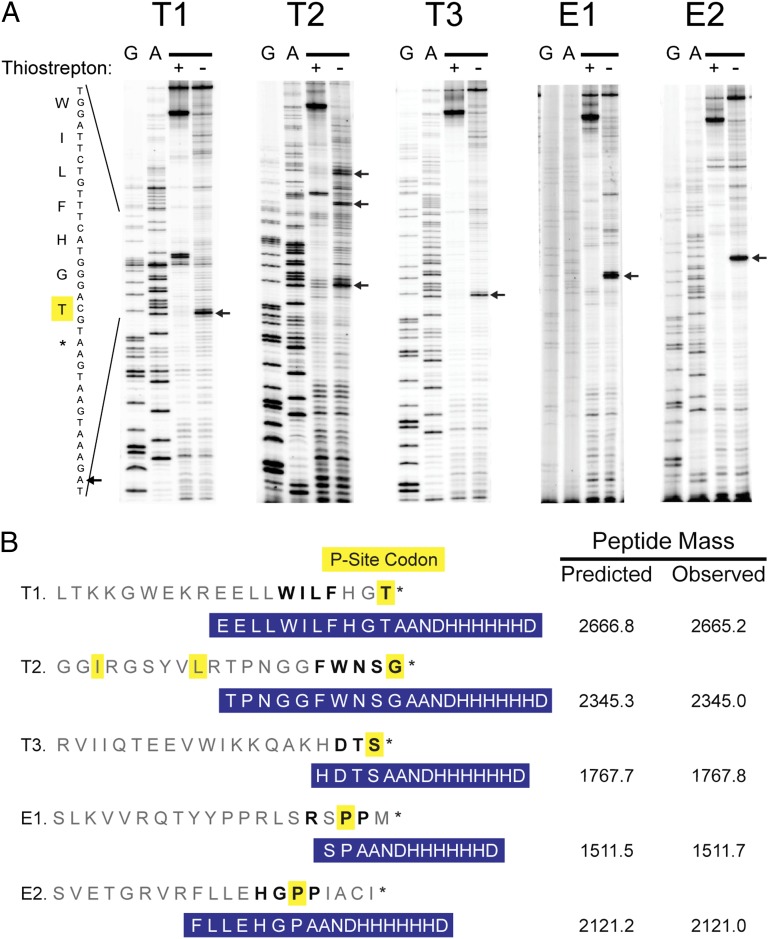
Fig. 2.
Stalling motifs that block termination (T1–T3) or elongation (E1 and E2). (A) Stalling sites for five motifs were determined by toeprinting assays. Blocks in cDNA synthesis are marked with arrows. The antibiotic thiostrepton traps ribosomes in initiation complexes; bands seen in both treated and untreated lanes are reverse transcriptase artifacts. (B) Key amino acids in each motif are written in bold. The codon labeled in yellow is positioned in the P site of the stalled ribosome. The site of stalling in vivo was confirmed by purifying tagged cI, digesting the protein with trypsin, and determining the mass of the C-terminal peptide (highlighted in blue) by MALDI-MS.