Fig. 1.
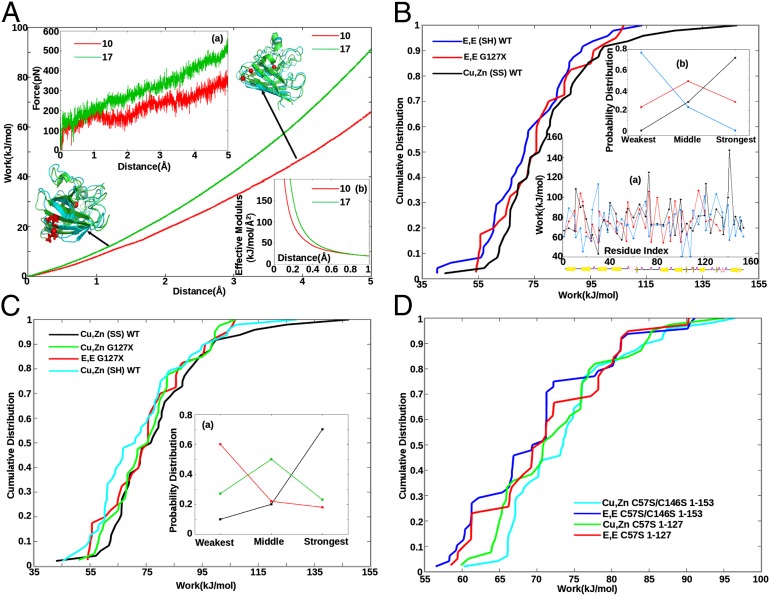
Force (A, a), work (A), and effective modulus (A, b) as a function of extension. Tethers are placed at the Cα atom closest to the center of mass of the SOD1 monomer (H46), and the Cα atom of either residues G10 (red) or I17 (green), in separate pulling assays. Ribbon representations of the protein are also shown; the tethering residue is shown in licorice rendering (in red) and the center Cα as a red sphere. The initial equilibrated (at 0 Å, green ribbon) and final (at 5 Å, blue ribbon) structures are aligned to each other by minimizing RMSD. (B, a) Work profiles of Cu,Zn(SS) WT (black), E,E(SH) WT (blue), and E,E G127X SOD1 (red) vs. sequence index. Secondary structure schematic is shown underneath. (B) Cumulative distributions of the work values in B, a. E,E G127X is more stable than full-length E,E (SH) SOD1 (P = 9e-7). (B, b) Fraction of the 48 incidences that each variant had either the weakest, strongest, or middle work value—e.g., E,E(SH) SOD1 is weakest 80% of the time and is never the strongest variant. (C) Cumulative work distributions for Cu,Zn (SS) WT (black), Cu,Zn G127X (green), E,E G127X (red), and Cu,Zn (SH) WT (cyan). Cu,Zn G127X is destabilized with respect to full-length Cu,Zn (SS) WT (P = 6.2e-8). (C, a) Same analysis as B, b for the variants Cu,Zn(SS) WT, Cu,Zn G127X, and E,E G127X . (D) Cumulative distributions for serine mutant SOD1 variants demonstrate that C-terminal truncation stabilizes the apo form but destabilizes the holo form (Results).