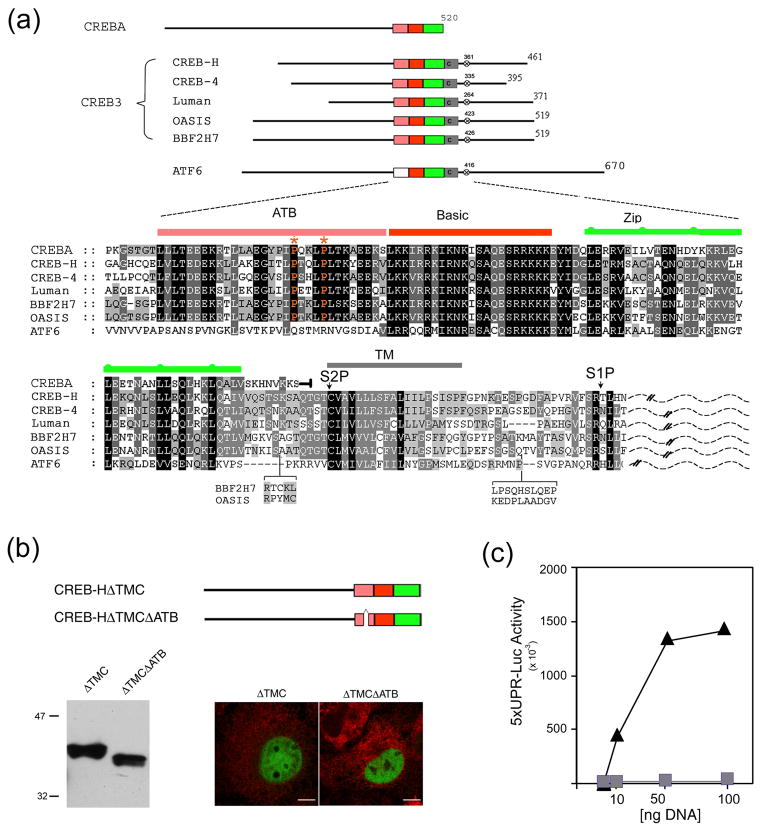
Figure 1. Requirement for the ATB domain in CREB-HΔTMC transcriptional activation.
(a) Organisation of the human CREB3 family members in relation to CREBA and ATF6, showing the ATB domain (pink), the basic region (red), the leucine zipper (green), the transmembrane (TM) domain (grey) and a conserved S1P motif (x). Primary sequence similarity within this central region is shown below. Alignment was performed using ClustalX and annotated with Genedoc. Shading reflects the degree of conservation weighted for chemical similarity. In CREBA, the sequence terminates immediately after the leucine zipper, whereas, in the CREB3 family and in ATF6, homology extends to the TM domain and S1P site. Lumenal sequences C-terminal to the TM domain are indicated by a dashed line. Note the absence of the ATB domain in ATF6, but its virtually complete conservation in CREBA. Optimal alignment based on sequences from additional species is discussed further (see Figure 9). (b) Illustration of CREB-HΔTMC and CREB-HΔTMCΔATB, which lacks 27 residues within the ATB region. COS cells were transfected with the appropriate vectors (1 μg) and expression levels assayed by Western blotting or by immunofluorescence using an antibody to the SV5 epitope tag (CREB-HΔTMC and CREB-HΔTMCΔATB, green channel; calreticulin (an ER marker), red channel). Scale bar, 10 μM. (c) Cells were transfected in triplicate with the target vector 5XATF6-GL3 (1 μg) containing a basal promoter with five copies of the UPR element, and increasing amounts (10 ng, 50 ng or 100 ng) of the expression vectors for CREB-HΔTMC (triangles) or CREB-HΔTMCΔATB (squares). Cells were harvested 24 h post-transfection and luciferase activation was measured.