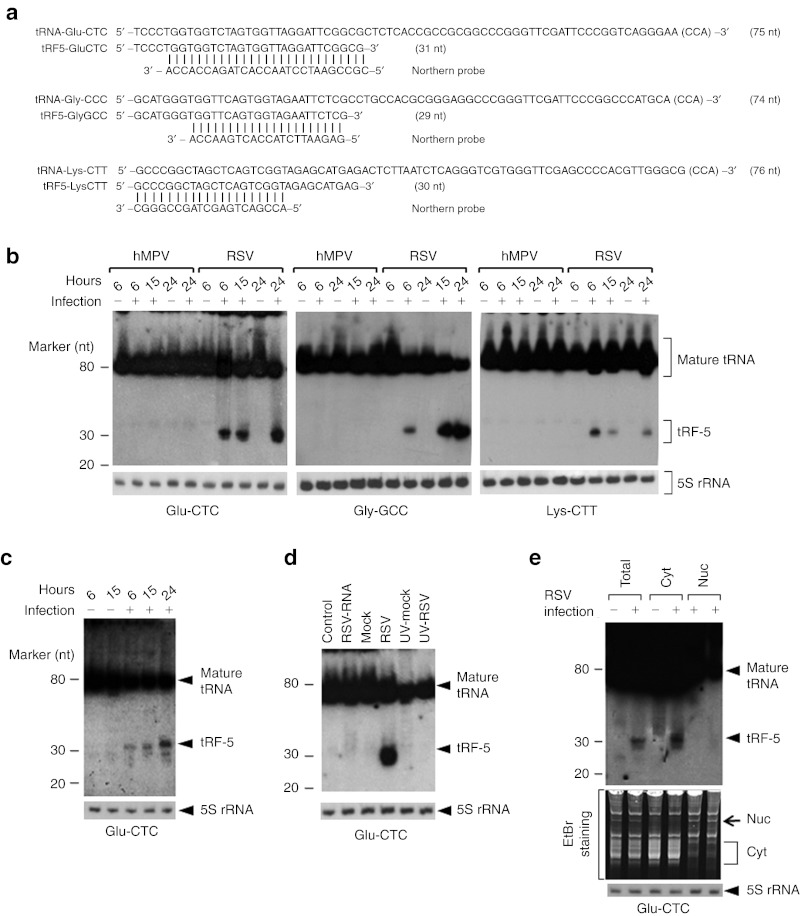
Figure 3.
Experimental validation and characterization of tRF-5s. (a) Sequence alignment of tRF-5s with their parental mature tRNAs and northern probes. The length of each sequence is indicated in parentheses. CCA sequence that is post-transcriptionally added to the 3′-end of tRNA is indicated in brackets. (b) Total RNA from indicated treatments in A549 cells was loaded to a denaturing polyacrylamide gel for northern hybridization using probes indicated in a; 5S rRNA is shown for equal loading. The positions of tRF-5 and mature tRNA are indicated on the right; molecular size markers are indicated on the left. The blot was exposed for ≤8 hours. Data are representative of 2–3 independent experiments. (c) Primary cultured SAE cells were mock- or RSV-infected at MOI of 3 for various lengths of time as indicated. All other experimental conditions were similar to those described in b, except that 4 µg of total RNAs were used for northern hybridization. (d) A549 cells were transfected with purified viral RNAs (lane 2), or infected with naive or UV-inactivated RSV for 24 hours (lanes 4 and 6). Cells treated with Lipofectamine 2000 (lane 1) or mock-infected (lanes 3 and 5) were used as negative controls. Northern hybridization was done as described in b. (e) Total, nuclear, or cytoplasmic RNAs from mock- or RSV-infected A549 cells were subjected to northern hybridization. All other descriptions are the same as in b. Data are representative of 2–3 independent experiments. EtBr, ethidium bromide; hMPV, human metapneumovirus; MOI, multiplicity of infection; nt, nucleotide; rRNA, ribosomal RNA; RSV, respiratory syncytial virus; SAE, small alveolar epithelial cell; tRF, tRNA-derived RNA fragment; tRNA, transfer RNA; UV, ultraviolet.