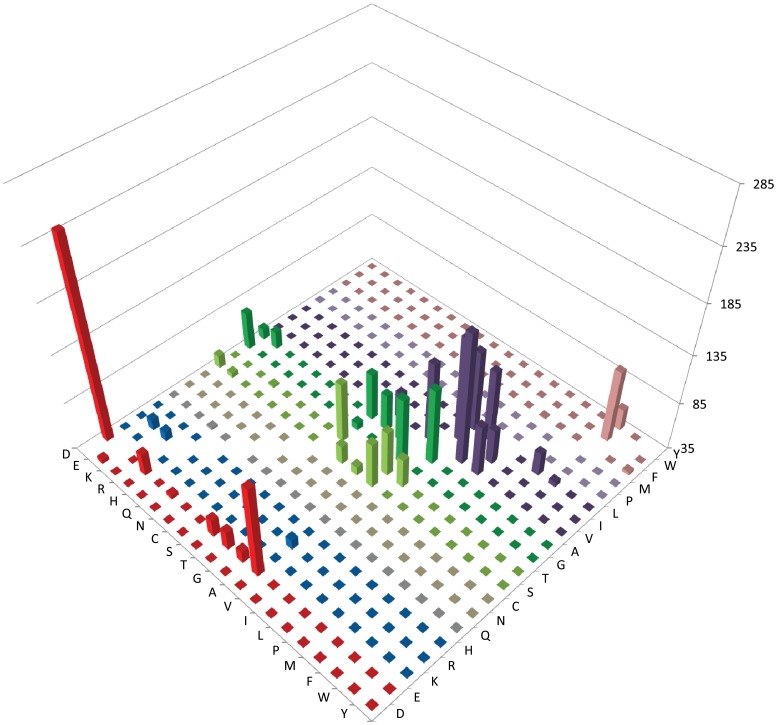
Figure 1. Amino acid substitution matrix of selected core haloarchaeal orthologous proteins for invariant residues in mesophilic versus cold-adapted H. lacusprofundi.
Amino acids conserved in 604 protein families in 12 mesophilic sequences are indicated on the right-axis, H. lacusprofundi amino acids are on left-axis, and the number of amino acid substitutions on the vertical axis, with the floor placed at 35 amino acids, to emphasize higher peaks.