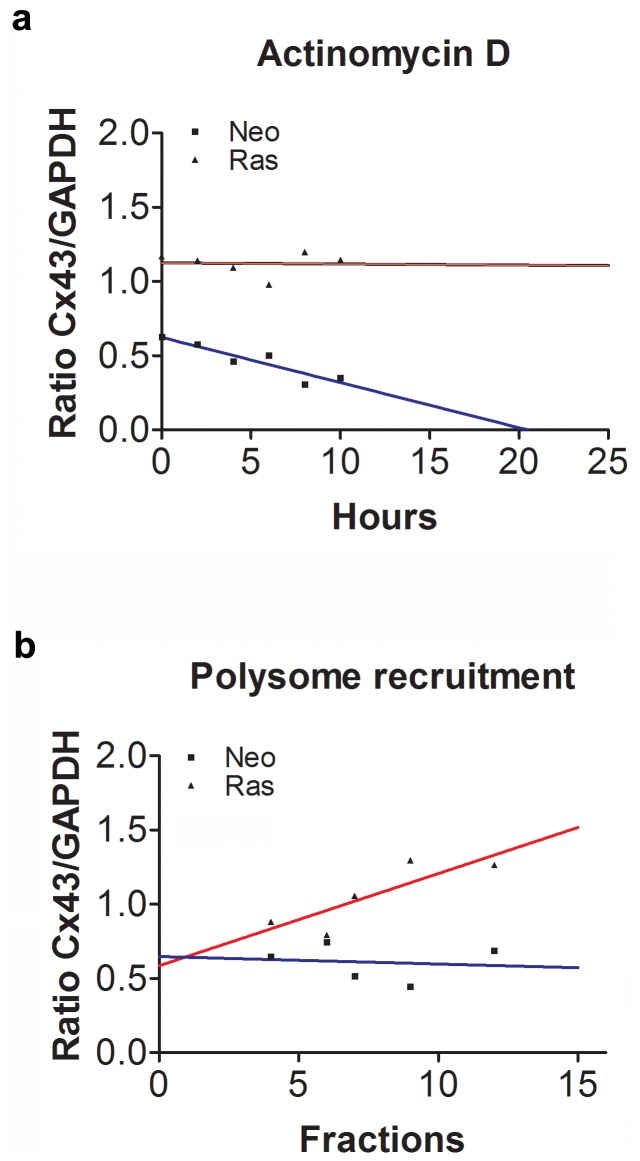
Figure 1. H-Ras regulates Connexin43 expression at post-transcriptional levels.
a) RNA stability measured by the rate of Cx43 mRNA decay following inhibition of de novo transcription by actinomycin D. NIH-3T3Neo and NIH-3T3Ras cells were treated with actinomycin D (15 µg/ml), harvested at various time points and Cx43 and GAPDH mRNA levels were determined by RT−PCR. The values of Cx43/GAPDH mRNAs ratios are reported over the duration of treatment. The lines are based on a linear regression and indicate that there is a significant slope deviation from the zero for Neo (r = 0.84; P = 0.0095) versus Ras (r = 0.0012; P = 0.94). b) Translation efficiency of the Cx43 mRNA. Polysomal and subpolysomal fractions were extracted from NIH-3T3Neo (Neo) and NIH-3T3Ras (Ras) cells using a sucrose density gradient and the relative levels of Cx43 and GAPDH mRNAs in these fractions were determined by RT-PCR. The results are reported as a Cx43/GAPDH mRNA ratio in different fractions. The lines are based on a linear regression and indicate that there is a significant slope deviation from the zero for Ras (r = 0.014; P = 0.84) versus Neo (r = 0.71; P = 0.07).