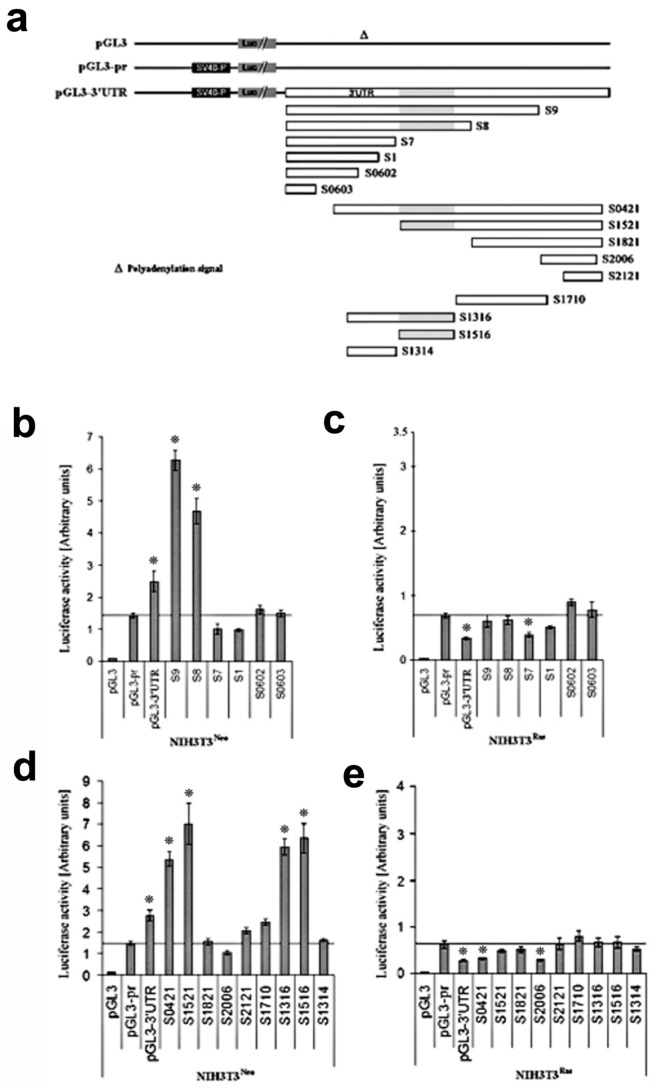
Figure 3. Characterization of the Cx43 mRNA 3′UTR regulatory regions in NIH3T3Neo and NIH3T3Ras by luciferase assay.
a) Localization of the different Cx43 3′UTR constructs used in this work. All constructs contain the SV40 promoter (SV40-P) and the Luciferase coding region in addition to the pGL3 polyadenylation signal after the 3′UTR segments (not shown in graphics). The position of a putative polyadenylation signal is indicated. The S1516 region is shown in light grey. b, c) Luciferase assay of the first set of 3′UTR constructs used to transfect NIH3T3Neo and NIH3T3Ras cells respectively. d, e) Luciferase assay of the second set of 3′UTR constructs used to transfect NIH3T3Neo and NIH3T3Ras cells respectively. The firefly luciferase activities were reported to the Renilla luciferase control values as explained in “Materials and Methods”. The experiments were performed at least three times in quadruplicates (*p<0.05, reported to the pGL3-pr construct).