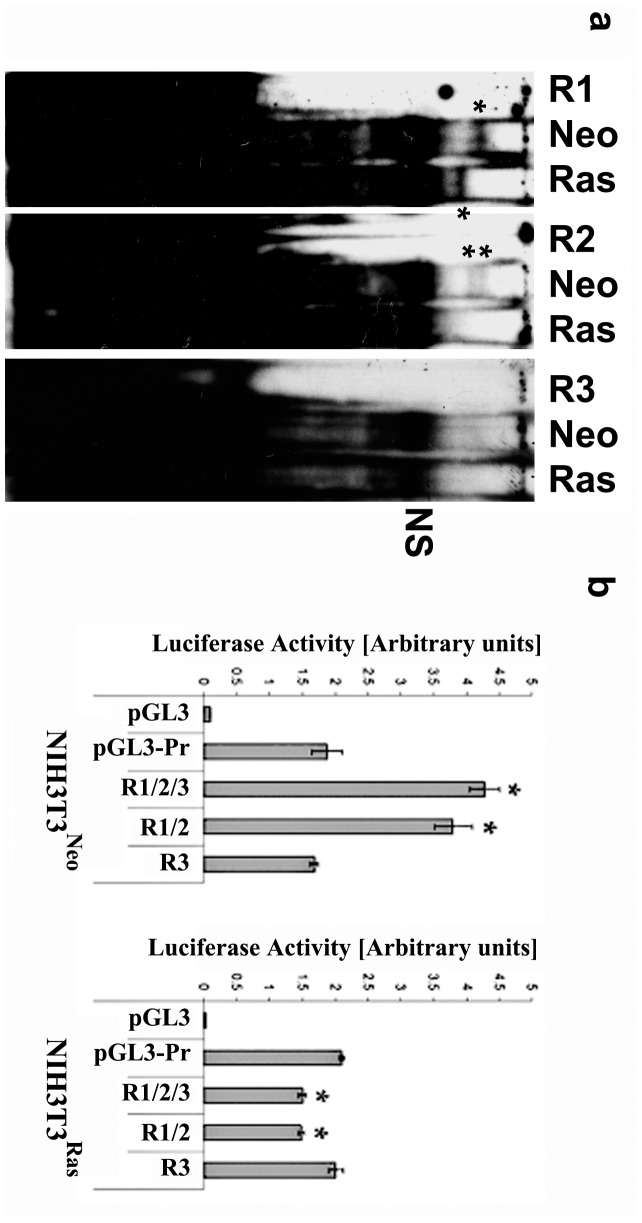
Figure 6. Differential binding of trans-acting factors to regions of the 3′UTR S1516.
a) REMSA using three different riboprobes (R1, R2 and R3) spanning the S1516 region. Binding reactions were performed with the riboprobes as explained in “Materials & Methods” in the presence of protein extracts from NIH3T3Neo (Neo) and NIH3T3Ras (Ras) cells. RNA/proteins complexes are indicated by stars. N.S.: non-specific binding. b) Luciferase assay using the NIH3T3Neo and NIH3T3Ras cells, transfected with Cx43 mRNA 3′UTR constructs corresponding to the three riboprobes R1, R2 and R3 (i.e. full length S1516 element, R1/2/3), riboprobes 1 and 2 (R1/2) or riboprobe 3 (R3). The experiments were performed at least three times in quadruplicates (*p<0.05). The experiments were performed at least three times in quadruplicates (*p<0.05).