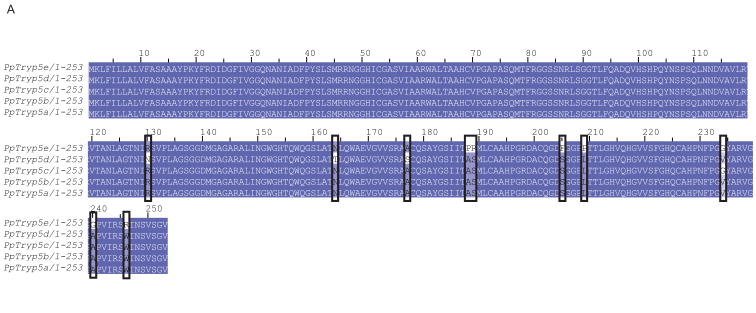
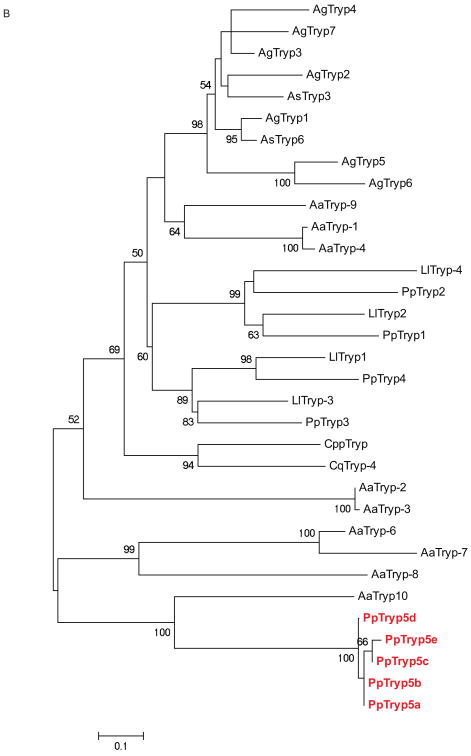
Figure 4. Ph. papatasi novel trypsin sequences.
(A) Novel Ph. papatasi multiple sequence alignment; conserved residues are represented by a darker shading while mismatches between the five sequences are indicated by boxes. Full sequence alignment is available in supplemental materials (S5). (B) Phylogenetic analysis of trypsin amino acid sequences from Ph. papatasi (Pp: PpTry5a-e (JP544502, JP542407, JP540627, JP554453, JP544448), AAM96940.1, AAM96941.1, AAM96942.1, AAM96943.1), Lu. longipalpis (Ll: ABM26904.1, ABM26905.1, ABV60308.1, ABV60300.1), An. gambiae (Ag: CAA80512.1, CAA79328.1, CAA80517.1, CAA80515.1, CAA80514.1, CAA80513.1, CAA80516.1), An. stephensi (As: AAB66878.1, AAA97479.1), Ae. aegypti (Aa: EAT40684.1, EAT42808.1, EAT36350.1, EAT34033.1, EAT42007.1, EAT42008.1, EAT42004.1, EAT37859.1), Cu. quinquefasciatus (Cq: EDS34988.1) and Cu. pipiens palens (Cpp: AAK67462.1). The WAG substitution model was used with variable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. The novel Ph. papatasi trypsin sequences (PpTryp5a-e) are indicated in bold.