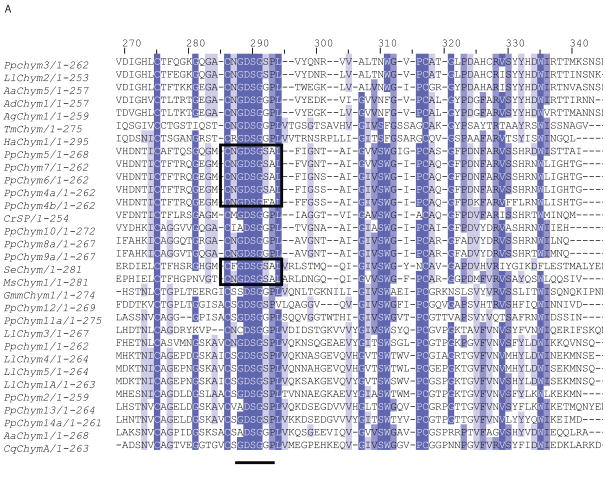
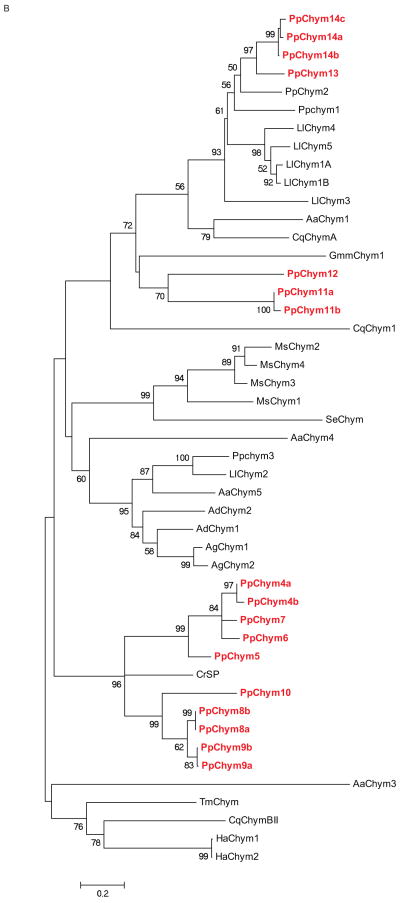
Figure 5. Ph. papatasi novel chymotrypsin sequences.
(A) Multiple sequence alignment fragment depicting the chymotrypsin binding pocket, where the pockets with the P-to-A substitution are boxed; the residue concordance at each position in indicated by the different degree of shading at that position. Full sequence alignment is available in supplemental materials (S6). (B) Phylogenetic analysis of known and novel chymotrypsins from Ph. papatasi (Pp: PpChym4-14 (JP546634, JP554565, JP551370, JP547341, JP554731, JP549601, JP540150, JP554746, JP554644, JP540516, JP543908, JP549141, JP547864, JP543702, JP542007, JP548909, JP549999), AAM96938.1, AAM96939.1, ABV44728.1), Lu. longipalpis (Ll: ABV60294.1, ABV60293.1, ABV60309.1, ABV60291.1, ABV60292.1, ABV60301.1), An. gambiae (Ag: CAA83568, CAA83567), An. darlingi (Ad: ADD17493.1, ADD17494.1), Ae. aegypti (Aa: XP_001663061.1, EAT32679.1, EAT38422.1, AAL93243.1), Cu. quinquefasciatus (Cq: XP_001846630.1, XP_001865429.1, XP_0011863473.1), Ch. riparius (Cr: ACF19792.1), Gl. morisitans morisitans (Gmm:ADD18377.1), He. armigera (Ha: ADI32883.1, ADI32881.1), Ma. sexta (Ms: CAL92020.1, CAM84317.1, CAM84318.1, CAM84319.1), Sp. exigua (Se: AAX35812.1) and Te. molitor (Tm: ABC88746.1). The WAG substitution model was used with variable and invariable positions and a bootstrap value of 1000 (only those above 50 are represented on the trees). The scale represents the rate of amino acid substitution per site. The novel Ph. papatasi chymotrypsin sequences are in bold.