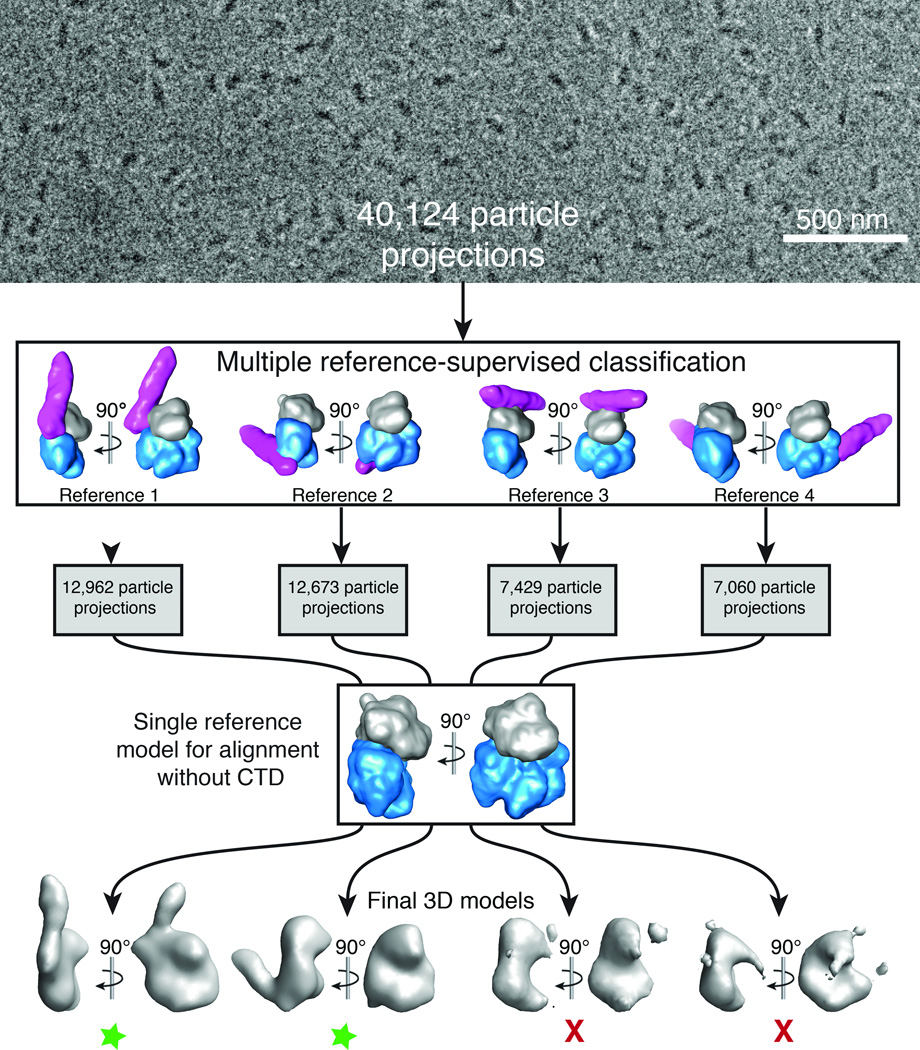
Figure 3.
Cryo-EM projection classification and 3D reconstruction scheme. 40,124 particle projections were subjected to multiple 3D reference-supervised classification against volumes of four Gαq–PLCβ3 core complexes observed in the crystal lattice. The density corresponding to Gαq is shown in gray, the PLCβ3 core in blue, and the distal CTD in purple. Each group of particles was used to generate a unique 3D reconstruction using the Gαq–PLCβ3 structure, omitting the distal CTD, as the initial reference. Two final 3D reconstructions were of good quality as judged by the definition of the Gαq–PLCβ3 core complex (green stars). In the first model the distal CTD is docked with the N-terminal helix of Gαq, and in the second with the hydrophobic ridge of the catalytic core of PLCβ3. The third represents a contact between the Dα3-Dα4 loop of the distal CTD and the second EF hand (Eα2 and Fα2 helices) of the catalytic core. The fourth model represents an interaction between the distal CTD and the Ras-like domain of Gαq (Supplementary Fig. 2d). Based on functional analysis (Table 2) and failure to obtain reliable reconstructions, the last two interactions (models 3 and 4) likely represent simple lattice contacts. A representative cryo-EM image of purified Gαq–PLCβ3 complexes is shown at the top of the figure.