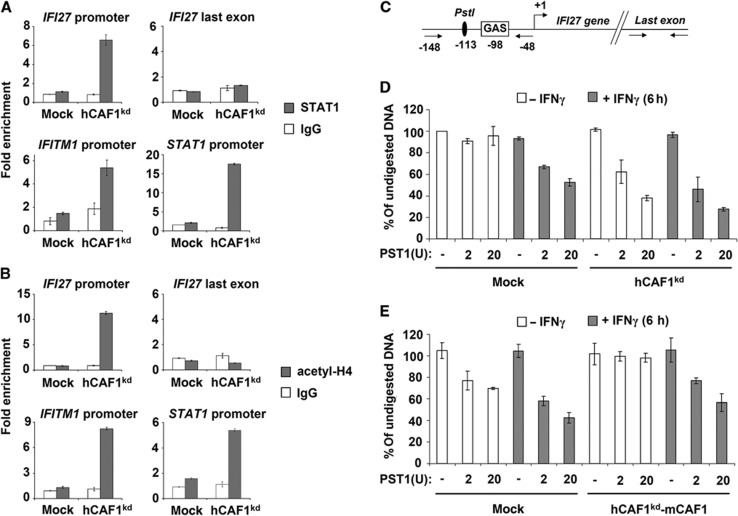
Figure 5.
Constitutive recruitment of STAT1 at a subset of STAT1-target promoters in hCAF1 knockdown cells. ChIP assays of untreated hCAF1kd and control cells were performed using antibodies anti-STAT1 (A) and anti-acetyl H4 (B). Enriched DNA fragments were quantified by qPCR using specific primers for the indicated promoters with respect to the input DNA and normalized to a reference locus (3′ downstream region of the GAPDH gene). Rabbit IgGs were used as a negative control. (C–E) hCAF1 affects chromatin accessibility. (C) Schematic representation of GAS, PST1 site and primer positions on IFI27 promoter. (D) hCAF1kd and control cells and (E) hCAF1kd transfected with empty pCIflag (mock) or with pCIflag-mCAF1 (hCAF1kd-mCAF1, rescued cells expressing mCAF1) were exposed or not to IFNγ for 6 h. Isolated nuclei were then treated with a limiting concentration of PST1 restriction enzyme, which cut GAS containing region in IFI27 promoter. DNA was then purified and the level of intact DNA was determined by qPCR using oligos flanking the GAS element or control region illustrated in (C). The experiments were performed in triplicate, expressed as mean values and are representative of at least three independent experiments. Standard deviations are shown.