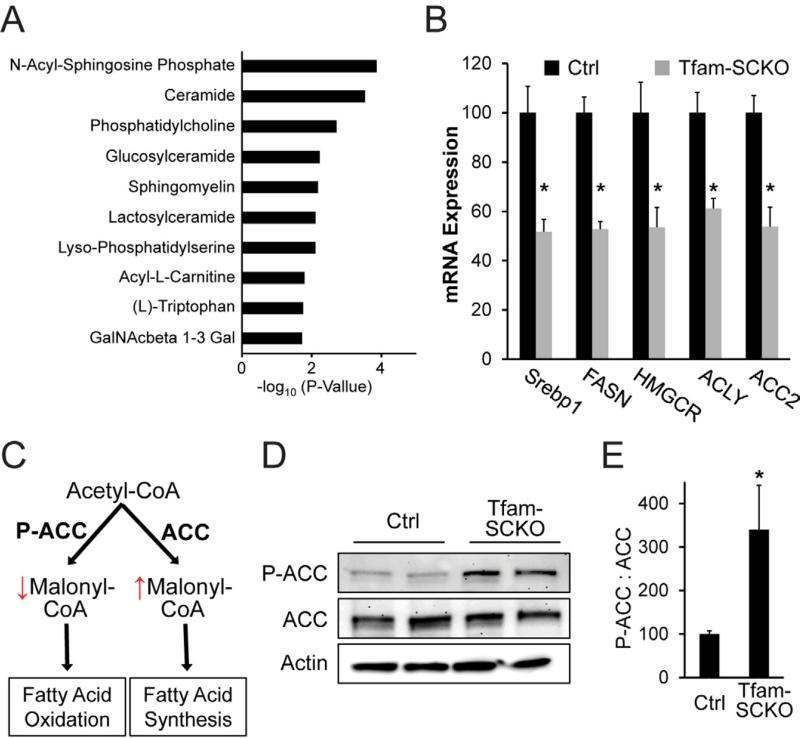
Figure 4. SC mitochondrial dysfunction causes a shift in lipid metabolism away from lipid biosynthesis and toward fatty acid oxidation.
(A) Differentially expressed mRNAs in 2-month-old Tfam-SCKO nerves as determined by microarray analysis are enriched for genes involved in lipid metabolism pathways. The 10 pathways with most significant enrichment among differentially expressed genes in Tfam-SCKO nerves are shown. (B) qRT-PCR analysis confirms that a number of lipid synthesis related enzymes are downregulated in 2-month-old Tfam-SCKO vs. Ctrl nerves. SREBP1, sterol regulatory element binding transcription factor 1; FASN, fatty acid synthase; HMGCR, 3-hydroxy-3-methylglutaryl-Coenzyme A reductase; ACLY, ATP citrate lyase; ACC2, acetyl-Coenzyme A carboxylase beta (primarily localized to mitochondria). N=5 mice per genotype. *P<0.05. (C) Diagram depicting the regulation by acetyl-coA carboxylase (ACC) of the balance between fatty acid synthesis vs. oxidation and how it is altered by ACC's phosphorylation status. (D, E) Immunoblot analysis (D) and quantification of band intensity (E) show increased phosphorylation of ACC in 2-month-old Tfam-SCKO nerves. Phosphorylation inhibits this central regulator of the balance between lipid synthesis vs. oxidation, indicating (together with gene expression results) a shift in lipid metabolism away from new lipid synthesis and towards increased lipid oxidation in SC following mitochondrial dysfunction. N=4 mice per genotype. *P<0.05.