Abstract
Molecular imaging is a rapidly advancing field that allows cancer biologists to look deeper into the complex inner workings of tumor cells, or whole tumors, in a non-invasive manner. In this review, we will summarize some recent advances that enable investigators to study various important biological processes in tumors in vivo. We will discuss novel imaging approaches that allow investigators to visualize and quantify molecular pathways, such as receptor tyrosine kinase activation, hypoxia signal transduction, apoptosis, and DNA double-strand breaks. Select examples of these applications will be discussed. Because of the limited scope of this review, we will only focus on natural reporters, such as bioluminescence and fluorescent proteins.
INTRODUCTION
In biomedical research, the ability to visualize a biological process continues to be a subject of investigation because it offers the most direct method to support or refute any scientific claim. Molecular biologists have exploited a wide variety of methods to visualize various molecular processes and signals. Often, surrogate reporters are used. One of the earliest such reporters is the bacterial enzyme β-galactosidase, which recognizes and cleaves the compound X-gal, and generates an intense blue product that can be identified and quantified (1). β-gal assays are widely used to quantify gene promoter activities. Another example is the green fluorescence protein (GFP); a jellyfish protein that was initially purified in the 1960s (2). The GFP gene was cloned in the 1980s (3) and is widely used to allow for direct visualization of transcriptional activities of promoters (4), or to locate a protein in live cells. Whole organisms, including animals and plants, have been engineered to carry the GFP gene so their cells and tissues could be traced easily (5). This widespread application of the GFP protein spurred many efforts to identify new variants that can fluoresce purple, cyan, yellow, red, etc., (6), and also stimulated efforts to identity additional fluorescent proteins suitable for a variety of applications in vivo. Another significant area of investigation for optical imaging is bioluminescence-based applications (7, 8), which offers new ways to study biology in vivo. In this review, we will summarize some of the more recent advances in bioluminescence- or fluorescent-protein-based approaches to study tumor biology in vivo.
LUCIFERASE-BASED BIOLUMINESCENCE APPROACHES
Bioluminescence, which is light production by living organisms, is very desirable as a reporter in biological applications. While it occurs in marine animals, microorganisms, and some terrestrial animals, the natural bioluminescence background in many organisms, including mammals, is zero. It is possible, therefore, to utilize these naturally occurring elements of bioluminescence to achieve exquisite sensitivity during imaging (9, 10). One of the most important bioluminescence reporter genes is the firefly luciferase gene (11). Firefly luciferase generates bioluminescence in a two-step reaction. The first step converts luciferin plus adenosine triphosphate (ATP) to luciferyl adenylate (with pyrophosphate). The second step requires oxygen to enable the forward reaction to occur with the reactant luciferyl adenylate, yielding the products oxy-luciferin, adenosine monophosphate (AMP), and light. The original chemical reaction without luciferase is extremely slow. Once the enzyme is introduced, the catalyzed reaction can be turned into a usable assay, and its photon product can be used effectively.
Advantages of the luciferase enzyme separate it from most other methods utilized for optical imaging in vivo. First, it does not require light to be activated. The bioluminescence effect is the end result of a chemical reaction. As long as oxygen, ATP, and the luciferin substrate are present in the environment, the highly-sensitive nature of the luciferase enzyme will continue to report signals. Second is the high signal-to-noise ratio; the lack of a natural bioluminescence background in most organisms gives an increased throughput. Third, with proper instrumentation (e.g., IVIS systems), in vivo signals can be quantified serially over time (9, 10). However, disadvantages of firefly luciferase include the need for substrate injection and the poor spatial resolution of bioluminescent imaging.
Traditionally, the luciferase assay has been mostly used in reporting transcriptional activity of promoters of interest in cells or mice. In recent years, many novel approaches to using the luciferase assay have been developed that allow one to visualize a wide array of molecular pathways. These assays provide very useful tools for radiation and cancer biology research. In the following sections of this review, we will summarize novel approaches to using firefly luciferase to image a variety of biological pathways.
A Split-Luciferase-Based Reporter for Monitoring Receptor Tyrosine Kinases
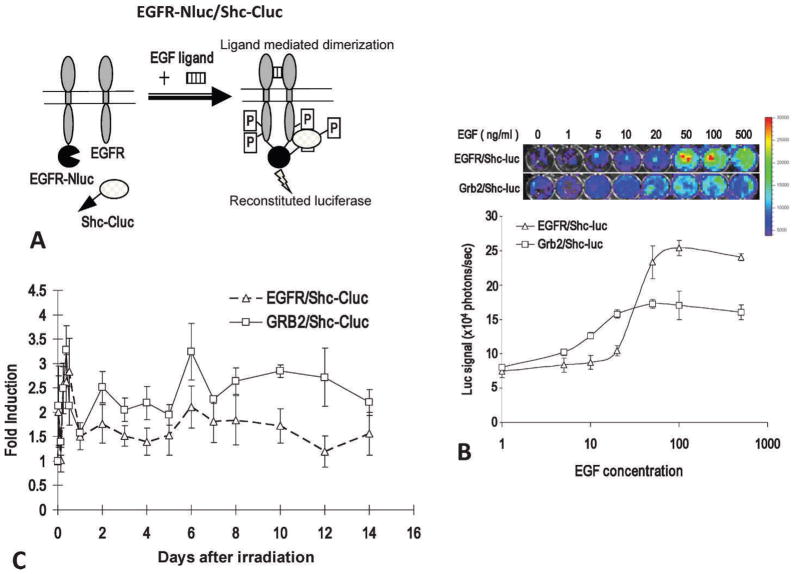
Epidermal growth factor (EGF) and its receptor, epidermal growth factor receptor (EGFR), represent a key cellular growth signal that plays an important role in the development and growth of many malignancies. Upon EGF binding, EGFR dimerizes to activate its tyrosine kinase activities. This leads to phosphorylation of itself and downstream targets, such as Grb2 and Shc. Phosphor-ylated Grb2 has been shown to physically associate with EGFR and Shc. Until recently, there had been no good method to noninvasively quantify the activity of EGFR activation. This has been remedied by Li et al. (12), who developed a method to measure EGFR activation based on the bifragment (or split) luciferase reconstitution system (13–16). In this system, two likely interacting protein partners are each fused to N- or C-terminal domains of the firefly luciferase gene, which does not have catalytic abilities of its own, and co-transduces into a cell of interest. When the two interacting proteins are brought together through any kind of signal activation, they also bring together the N- and C-terminal halves, which, reconstitutes the enzymatic abilities of luciferase. Li et al. fused EGFR and its downstream binding partners Grb2 and Shc to N- and C-terminal halves of firefly luciferase (Fig. 1A). They showed that pairs of EGFR-Nluc/Shc-Cluc, or Grb2-Nluc/Shc-Cluc, worked well in serving as surrogates of EGFR activation. Both show excellent EGF-induced signal activation in vitro (Fig. 1B), and radiation-induced EGFR in vivo (Fig. 1C). In addition, this system was used to observe hyperthermia-induced EGFR activation in tumor cells and the potential mechanisms involved (17), and to observe RTK activation for the PTEN-Akt pathway in tumor cells (19).
FIG. 1.
Imaging EGFR activation through the use of the split-luciferase system. Panel A: Graphic illustration of the principle of a split-luciferase EGFR-Shc activity reporter. Panel B: The dose-response curve for the EGFR/Shc-luc reporter and the Grb2/Shc-luc reporter. The top panel shows representative images of reporter-transduced cells (in 48-well plates) treated with different concentrations of EGF at 37°C 15 min and then imaged in the IVIS200 instrument, while the lower panel shows the quantitative dose-response of the reporter activation after EGF addition. The error bars represent standard deviations derived from 3 to 5 data points. Panel C: Radiotherapy-induced activation of EGFR/Shc-luc (broken lines) and Grb2/Shc-luc (solid lines) reporters in H322 xenograft tumors. After reporter-transduced H322 lung tumor cell (5 × 106) implantation (subcutaneous) and tumor formation (with diameters around 5 to 7 mm), the tumors were irradiated with X rays (6 Gy). The activities of the EGFR were then imaged.
A similar reporter system was developed to monitor the activities of the ErbB2 receptor, which belongs to the EGFR family and plays a key role in breast cancer development and treatment. With the aid of this reporter, it was shown that radiotherapy could significantly activate the ErbB2 reporter in xenograft tumors (18).
Observation of DNA Double-Strand Breaks in Live Cells Exposed to Ionizing Radiation
Another exciting advance is the ability to image DNA double-strand breaks in tumors exposed to ionizing radiation through bioluminescence imaging. DNA double-strand breaks (DSB) are the most important lesions generated by cellular exposure to ionizing radiation. Over the years, a large number of DNA DSB repair factors and their biological roles have been identified. However, DSB repair kinetics, especially those in vivo, are not well studied due to a lack of appropriate tools. In a recently published study, Li et al. developed a novel split-luciferase-based system to detect DSBs in live cells and tumors in a quantitative and non-invasive manner (20). The investigators took advantage of the fact that, right after the generation of DSBs in a cell, histone H2AX is rapidly recruited and phosphorylated at the sites of DSBs. Accumulation of phosphorylated H2AX (γ-H2AX) further recruits a number of additional proteins involved in DNA repair, checkpoint activation, and chromatin modification. A key protein that physically associates with phosphorylated H2AX is the protein mediator of DNA damage checkpoint, or MDC1. The main function of MDC1 is its collaboration with H2AX to recruit factors responsible for checkpoint activation or DNA damage repair (21, 22). More importantly, MDC1 was shown to associate with H2AX in a phosphorylation-dependent manner (23). To establish a reporter for DNA DSBs, Li et al. used γ-H2AX as a surrogate marker. To measure the amount of γ-H2AX, the investigators fused H2AX and MDC1 genes to N- and C-terminal halves of the firefly luciferase gene; H2AX was fused to the N-terminal half while the BRCT domain of MDC1 was fused to the C-terminal half. Since the BRCT domain is located in the carboxyl domain of MDC1 and has been shown to interact with phosphorylated ser139 in the H2AX, the investigators reasoned that, with the fusion reporter proteins expressed in the target cells, DNA damage, such as DSBs, would lead to the phosphorylation of H2AX and its interaction with MDC1. Such an interaction was likely to bring together the N- and C-terminal halves of the luc2 enzyme, which might lead to the reconstitution of luc2.
The reporter was activated in irradiated lung cancer cells in a very robust manner with a clear dose-dependent response. Reporter activities were consistent with Western blot analysis of γ-H2AX. Most interestingly, the investigators observed that reporter activities experienced second peaks around days 5 and 7 in cells that have been exposed to 6 Gy of irradiation. This finding had not been reported before due to the difficulty of monitoring and quantifying H2AX foci for an extended period of time. Most importantly, when tumors were established from the reporter-transduced cells, radiation-induced γ-H2AX was readily observable through bioluminescence imaging. In fact, activities of the reporters could be serially imaged for over 2 weeks. It is also possible that mice with the reporters engineered as transgenes could be observed for their whole lifetime for DSB induction through bioluminescence imaging.
Monitoring Apoptosis and Caspase Activation in Tumors Exposed to Ionizing Radiation
Another interesting and novel application of firefly luciferase is using it to monitor caspase activation in cancer cells exposed to therapeutic agents. Huang et al. used a novel reporter system to detect caspase 3 activation in irradiated tumors (24). In this reporter system (Fig. 2A), a luciferase-GFP fusion protein is linked to a polyubiquitin domain. In between the two moieties, a caspase 3 cleavage site (DEVD) is inserted. The assumption is that, when caspase 3 is inactive, the reporter proteins will be recognized by proteasomes and degraded immediately, because the polyubiquitin domain serves as tag for protein destruction by proteasomes. However, when caspase 3 is active, the polyubiquitin domain will be cleaved off the reporter protein, leading to enhanced GFP and luciferase signals, because these reporter proteins are no longer subject to direct proteasome recognition and degradation. Tumors were established from caspase 3-transduced 4T1 cells and were exposed to radiotherapy. Data indicate that significant caspase 3 activation occurred after radiotherapy in the 4T1 tumors (Fig. 2B). In fact, caspase 3 activation was observed to be over 30-fold. These data show that the proteasome-based bioluminescence caspase 3 reporter has a much wider dynamic range than previously reported, and compares favorably to another bioluminescence-based reporter of caspase 3 (25). In addition, the proteasome-based approach is suitable for the observation of any protease where the cleavage sequence is defined.
FIG. 2.
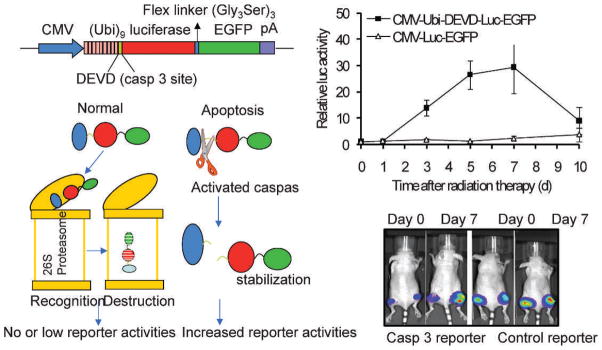
Imaging caspase activation during radiotherapy of cancer. Caspase 3 activation in 4T1 tumors as detected by a caspase 3 reporter. Left panels depict the structure of a proteasome-based caspase 3 reporter (top left) and its principle of action (lower left). Right panels showed caspase 3 activities in 4T1 tumors transduced with the control, as well as caspase reporter genes. The difference between the control and caspase 3 reporter groups are significant at days 3, 5, and 7 (P < 0.01, n = 5, t test). Error bars, SEM.
Monitoring Activation of Hypoxia-Inducible Factor Though the Use of a Unique Bioluminescence Fusion Reporter
Firefly luciferase is a reporter that can also allow an investigator to accurately monitor activities of hypoxia-inducible factor 1 (HIF-1α) (26), which plays a critical role in mammalian cellular response to low oxygen tension. This has been shown to have a key function in regulating the expression of vascular endothelial growth factor (VEGF) in normal, as well as malignant, tissues (27, 28). Therefore, there has been strong interest in the activities of HIF-1. To monitor the activities of HIF-1α, Li et al. exploited the fact that the activities of HIF-1α largely depend on its overall level in a cell, which is regulated by the ubiquitin-proteasome system in an oxygen-dependent manner. A 200 amino acid long oxygen-dependent domain (ODD) confers the oxygen sensitivity (29, 30). Under normoxic conditions, two proline residues in the ODD domain are hydroxylated by prolyl hydroxylase, which makes HIF-1α recognizable by the E3 ubiquitin ligase VHL. Recognition of HIF-1α by VHL leads to the ubiquitylation of the former and leads to its rapid degradation by the proteasome system. Li et al. fused the ODD with firefly luciferase to derive the reporter gene ODD-luc (31). They showed that, when introduced into human and mouse tumor cells, ODD-luc can recapitulate the level and transcriptional activities of HIF-1α much more accurately than a promoter-based system. Previously, an artificial HIF-1 promoter based on HIF-1 binding elements has shown some HIF-1 responsiveness, but also suffers from high background levels that greatly narrow its dynamic range. With this new reporter, the investigators were able to monitor HIF-1 activity in irradiated tumors in mice for an extended period of time and determine that, during radiotherapy, an oxygen-independent mechanism of HIF-1 regulation based on nitric oxide and generated by induced nitric oxide synthase (iNOS) was responsible for radiation-induced HIF-1α stabilization of HIF-1 activation and tumor resistance.
NEAR-INFRARED FLUORESCENT PROTEINS FOR IN VIVO APPLICATION
Since the 1990s, fluorescent proteins have gained popularity as the most widely used reporter system for molecular biology applications, such as promoter assays and protein localization tracers. The advantages of fluorescent proteins are obvious; no substrates for the proteins are needed. The presence of an external exciting wavelength is enough for the proteins to emit fluorescence (6). In addition, spatial resolution is exquisitely high. Therefore, they are especially suited for in vitro live cell applications, where spatial resolution is important. However, the use of fluoroprotein-based optical imaging in vivo has lagged behind bioluminescence-based approaches, since the former was faced with the challenge of strong tissue absorption of most of the light in the visible spectrum. Most fluorescent proteins, such as GFP, CFP, and YFP, emit fluorescence at wavelengths close to the green spectrum, which are especially prone to tissue absorption. In addition, autofluorescence from the tissues interferes with the fluorescent signals towards the green spectrum. To overcome this problem, investigators have searched for various fluorescent proteins with emission wavelengths in the red spectrum. One of the first red fluorescent proteins is from the coral Discosoma (32). It is normally a tetrameric protein, which limits its applications. However, it was determined that a monomeric version of the protein is functional, which makes the protein amenable for fusion with other proteins (33). Also, use of the monomeric protein shifted the excitation/emission wavelengths from 558/583 to 584/607 nm. Many other red-shifted GFP-like proteins have since been generated. However, even the most red-shifted proteins still show emission spectra outside the optimal spectral range for in vivo application, which should lie within 650 to 900 nm, a range labeled as the near-infrared optical window (NIRW), which has the lowest tissue absorbance for hemoglobin, water, lipids, and light scattering. [For additional information, see the review by Palmer et al. in this same issue (34).]
Significant progress continues to occur in this area. Shu et al. reported the successful engineering of an infrared fluorescence protein from the phytochrome of radiation-resistant extremophile bacterium Deinococcus radiodurans, which the authors dubbed as IFP1.4 (35). IFP1.4 incorporates biliverdin as its chromophore; it has excitation and emission wavelengths of 684 and 708 nm, respectively. Because biliverdin is ubiquitous as the initial intermediate in heme catabolism in mammals and is by itself non-fluorescent, IFP1.4 should have great promise in in vivo applications. However, fluorescence properties of IFP1.4, such as effective brightness and photostability, still require improvement. Filonov et al. (36) recently engineered another near-infrared protein based on phytochrome RpBphP2; the photosynthetic bacterium Rohopseudomonas palustris. Similar to IFP1.4, it also can exploit endogenous biliverdin and has excitation and emission wavelengths of 690 and 713 nm. In addition, the new IRFP (infrared fluorescent protein) is significantly brighter and more stable, and exhibits a very high signal-to-noise background ratio in mice.
The advent of these new near-infrared proteins should open up new possibilities for fluorescence-based imaging in in vivo tumor biology in animal models. With these proteins, it is now possible to trace and monitor the development and growth of a small number of tumor cells with great spatial resolution. It also is possible to monitor the status of various biological pathways in a live tumor without invasive surgery or substrate injection.
CONCLUSION
Bioluminescence imaging is now a well-developed modality for the non-invasive visualization of tumor cells and molecular pathways within tumor cells; it will continue to improve with the availability of additional new and innovative reporter systems. In contrast, fluorescent protein-based systems, despite their widespread use in vitro, lag behind due to the lack of suitable near-infrared fluorescent proteins that can penetrate tissues effectively. Newly engineered phytochrome-based fluorescent proteins, such as IFP1.4 or IRFP, should significantly change this scenario. Fluorescent proteins have the advantage of offering greater spatial resolution without the need for substrate injections. It is almost certain that the application of either, or both, of these types of reporter systems will provide significant insights into molecular mechanisms of in vivo radiation and tumor biology in the future.
Acknowledgments
This work was supported in part by grants CA131408, CA136748, and CA155270 from the U.S. National Cancer Institute (Li), and grant NNX12AB88G (Li) from U.S. National Aeronautics and Space Administration Ground based Space Radiation Biology Research Program.
References
- 1.Fowler AV, Zabin I. The amino acid sequence of beta galactosidase. I. Isolation and composition of tryptic peptides. J Biol Chem. 1970;245:5032–5041. [PubMed] [Google Scholar]
- 2.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 3.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 4.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Meth. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 7.Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, Stevenson DK, Benaron DA. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 8.Negrin RS, Contag CH. In vivo imaging using bioluminescence: a tool for probing graft-versus-host disease. Nature Rev. 2006;6:484–490. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 9.Cao YA, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci USA. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweeney TJ, Mailander V, Tucker AA, Olomu AB, Zhang W, Cao Y, Negrin RS, Contag CH. Visualizing the kinetics of tumor-cell clearance in living animals. Proc Natl Acad Sci USA. 1999;96:12044–12049. doi: 10.1073/pnas.96.21.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould SJ, Subramani S. Firefly luciferase as a tool in molecular and cell biology. Analytical Biochem. 1988;175:5–13. doi: 10.1016/0003-2697(88)90353-3. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Li F, Huang Q, Frederick B, Bao S, Li CY. Noninvasive imaging and quantification of epidermal growth factor receptor kinase activation in vivo. Cancer Res. 2008;68:4990–4997. doi: 10.1158/0008-5472.CAN-07-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 14.Gross S, Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massoud TF, Paulmurugan R, De A, Ray P, Gambhir SS. Reporter gene imaging of protein-protein interactions in living subjects. Curr Opin Biotechnol. 2007;18:31–37. doi: 10.1016/j.copbio.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf F, Li W, Li F, Li CY. Non-invasive, quantitative monitoring of hyperthermia-induced EGFR activation in xenograft tumours. Int J Hyperthermia. 2011;27:427–434. doi: 10.3109/02656736.2011.566593. [DOI] [PubMed] [Google Scholar]
- 18.Wolf F, Li W, Li F, Li CY. Novel luciferase-based reporter system to monitor activation of ErbB2/Her2/neu pathway noninvasively during radiotherapy. Int J Radiat Oncol Biol Phys. 2011;79:233–238. doi: 10.1016/j.ijrobp.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, Holland EC, Ross BD, Rehemtulla A. Molecular imaging of Akt kinase activity. Nature Med. 2007;13:1114–1119. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Li F, Huang Q, Shen J, Wolf F, He Y, Liu X, Hu YA, Bedford JS, Li CY. Quantitative, noninvasive imaging of radiation-induced DNA double-strand breaks in vivo. Cancer Res. 2011;71:4130–4137. doi: 10.1158/0008-5472.CAN-10-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 22.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 23.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 24.Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, O’Sullivan B, He Z, Peng Y. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nature Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laxman B, Hall DE, Bhojani MS, Hamstra DA, Chenevert TL, Ross BD, Rehemtulla A. Noninvasive real-time imaging of apoptosis. Proc Natl Acad Sci USA. 2002;99:16551–16555. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci USA. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:S62–67. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 28.Semenza GL. Targeting HIF-1 for cancer therapy. Nature Rev. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 29.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFa targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 30.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 33.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer GM, Vishwanath K, Dewhirst MW. Application of optical imaging and spectroscopy to radiation biology. Radiat Res. 2012;177:000–000. doi: 10.1667/rr2531.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]