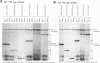Abstract
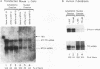
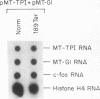
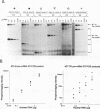
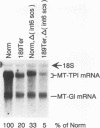
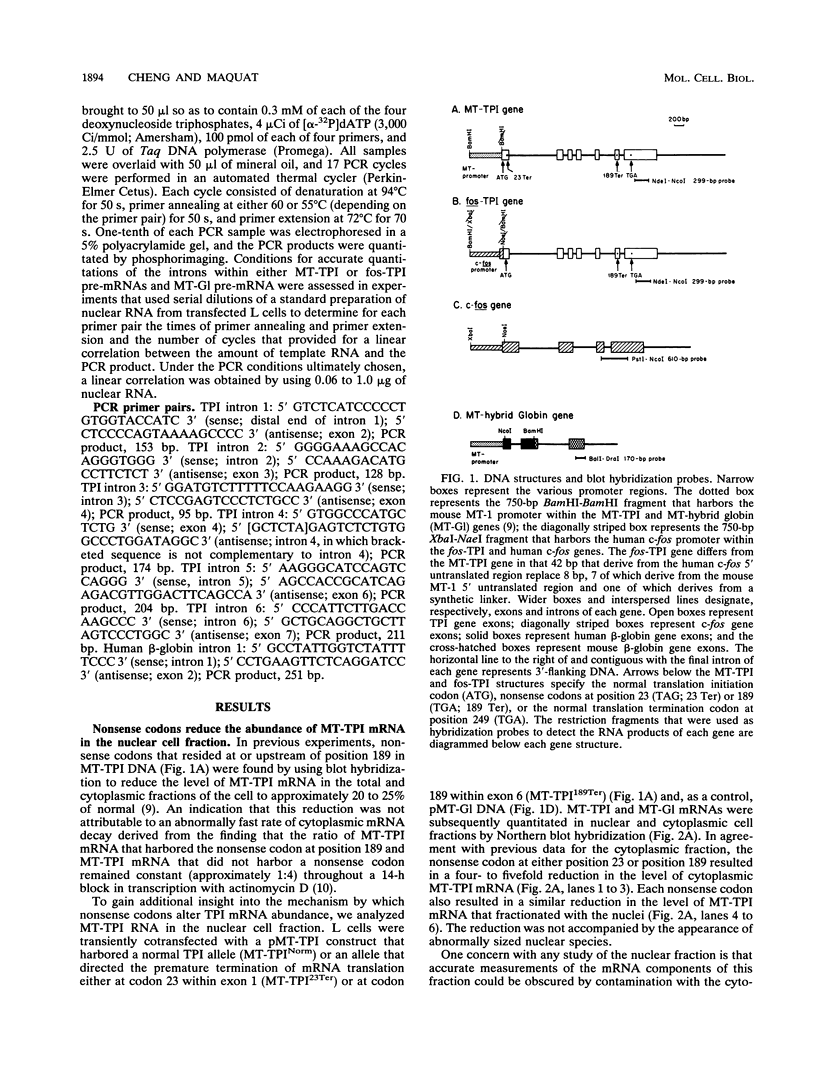
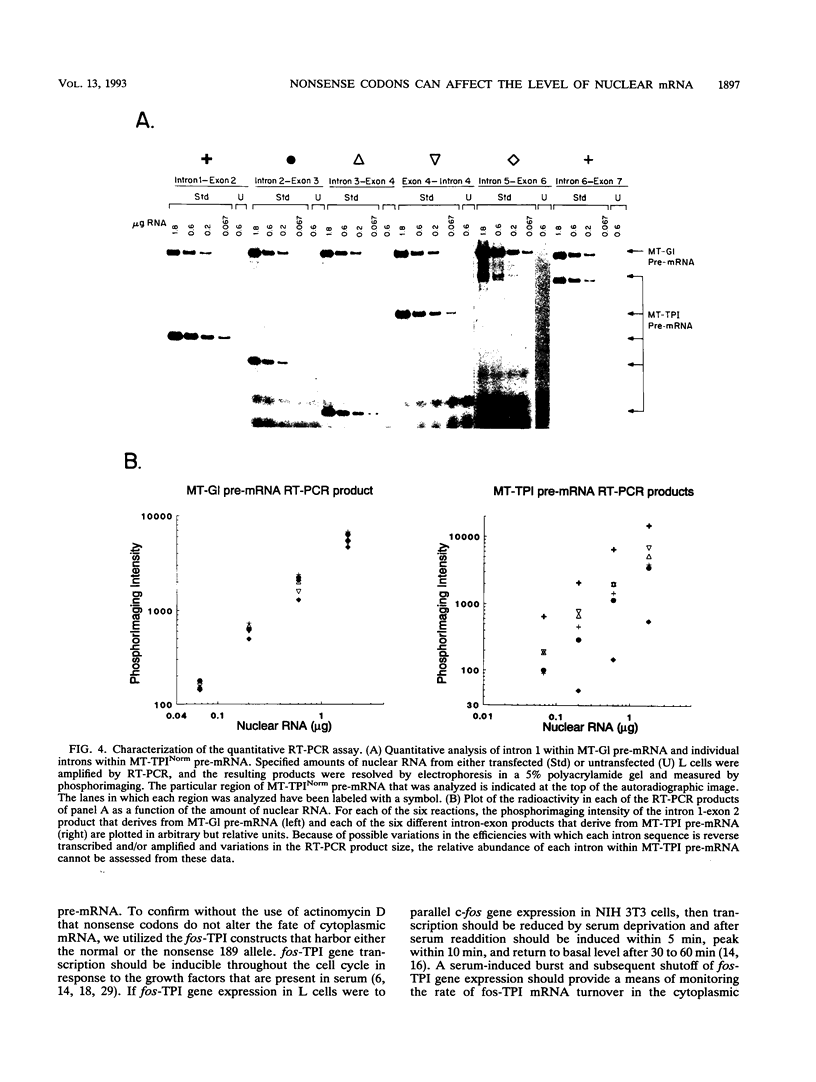
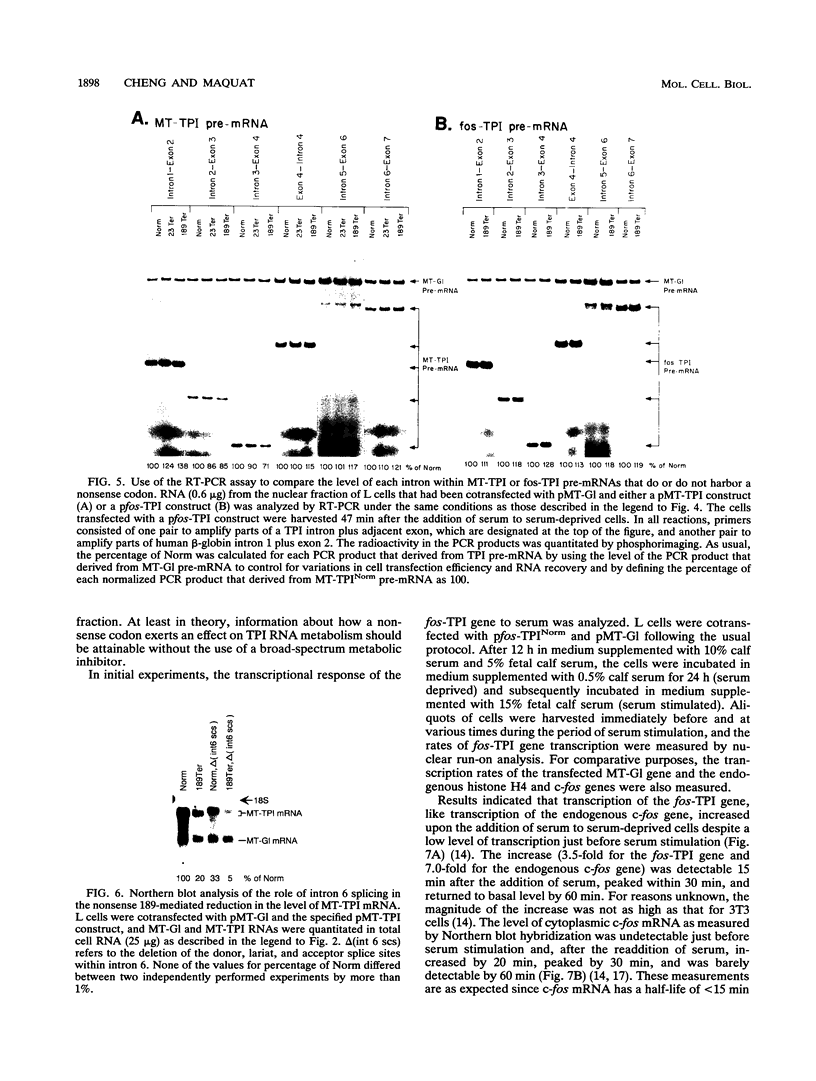
The abundance of the mRNA for human triosephosphate isomerase (TPI) is decreased to approximately 20% of normal by frameshift and nonsense mutations that cause translation to terminate at a nonsense codon within the first three-fourths of the reading frame. Results of previous studies inhibiting RNA synthesis with actinomycin D suggested that the decrease is not attributable to an increased rate of cytoplasmic mRNA decay. However, the step in TPI RNA metabolism that is altered was not defined, and the use of actinomycin D, in affecting all polymerase II-transcribed genes, could result in artifactual conclusions. In data presented here, the nonsense codon-mediated reduction in the level of TPI mRNA is shown to be characteristic of both nuclear and cytoplasmic fractions of the cell, indicating that the altered metabolic step is nucleus associated. Neither aberrancies in gene transcription nor aberrancies in RNA splicing appear to contribute to the reduction since there were no accompanying changes in the amount of nuclear run-on transcription, the level of any of the six introns in TPI pre-mRNA, or the size of processed mRNA in the nucleus. Deletion of all splice sites that reside downstream of a nonsense codon does not abrogate the reduction, indicating that the reduction takes place independently of the splicing of a downstream intron. Experiments that placed TPI gene expression under the control of the human c-fos promoter, which can be transiently activated by the addition of serum to serum-deprived cells, verified that there is no detectable effect of a nonsense codon on the turnover of cytoplasmic mRNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988 May;8(5):2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G. F., Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991 May;11(5):2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Beta-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2935–2939. doi: 10.1073/pnas.89.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Burckhardt J., Curran T., Müller R. Expression of c-fos in NIH3T3 cells is very low but inducible throughout the cell cycle. EMBO J. 1986 Apr;5(4):695–700. doi: 10.1002/j.1460-2075.1986.tb04269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso O., Bleecker G. C., Heintz N. Sequences controlling histone H4 mRNA abundance. EMBO J. 1987 Jun;6(6):1825–1831. doi: 10.1002/j.1460-2075.1987.tb02437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Grunberger D., Chasin L. A. Point mutation analysis in a mammalian gene: rapid preparation of total RNA, PCR amplification of cDNA, and Taq sequencing by a novel method. Biotechniques. 1989 May;7(5):494-6, 498-9. [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic M., Maquat L. E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990 Oct;10(10):5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Hermanowski A. L., Ziff E. B. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol. 1986 Apr;6(4):1050–1057. doi: 10.1128/mcb.6.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spector D. L. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991 Dec;5(12A):2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., Cooper J. A., Hunter T., Verma I. M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984 Dec 20;312(5996):711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H., Marselle L. M. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989 May 5;57(3):493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Leeds P., Peltz S. W., Jacobson A., Culbertson M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. K., Maquat L. E. Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5' termini. EMBO J. 1992 Sep;11(9):3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. K., Sigmund C. D., Gross K. W., Maquat L. E. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992 Mar;12(3):1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Chilcote R., Ryan P. M. Human triosephosphate isomerase cDNA and protein structure. Studies of triosephosphate isomerase deficiency in man. J Biol Chem. 1985 Mar 25;260(6):3748–3753. [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Mehlin H., Daneholt B., Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992 May 15;69(4):605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- Müller R., Bravo R., Burckhardt J., Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984 Dec 20;312(5996):716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT alpha 1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp R. A., Weintraub H. Ubiquitous MyoD transcription at the midblastula transition precedes induction-dependent MyoD expression in presumptive mesoderm of X. laevis. Cell. 1991 Jun 14;65(6):927–937. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- Senapathy P. Origin of eukaryotic introns: a hypothesis, based on codon distribution statistics in genes, and its implications. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2133–2137. doi: 10.1073/pnas.83.7.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Sierra F., Stein G., Stein J. Structure and in vitro transcription of a human H4 histone gene. Nucleic Acids Res. 1983 Oct 25;11(20):7069–7086. doi: 10.1093/nar/11.20.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Yen T. J., Gay D. A., Pachter J. S., Cleveland D. W. Autoregulated changes in stability of polyribosome-bound beta-tubulin mRNAs are specified by the first 13 translated nucleotides. Mol Cell Biol. 1988 Mar;8(3):1224–1235. doi: 10.1128/mcb.8.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. J., Machlin P. S., Cleveland D. W. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature. 1988 Aug 18;334(6183):580–585. doi: 10.1038/334580a0. [DOI] [PubMed] [Google Scholar]
- Zambetti G., Ramsey-Ewing A., Bortell R., Stein G., Stein J. Disruption of the cytoskeleton with cytochalasin D induces c-fos gene expression. Exp Cell Res. 1991 Jan;192(1):93–101. doi: 10.1016/0014-4827(91)90162-n. [DOI] [PubMed] [Google Scholar]