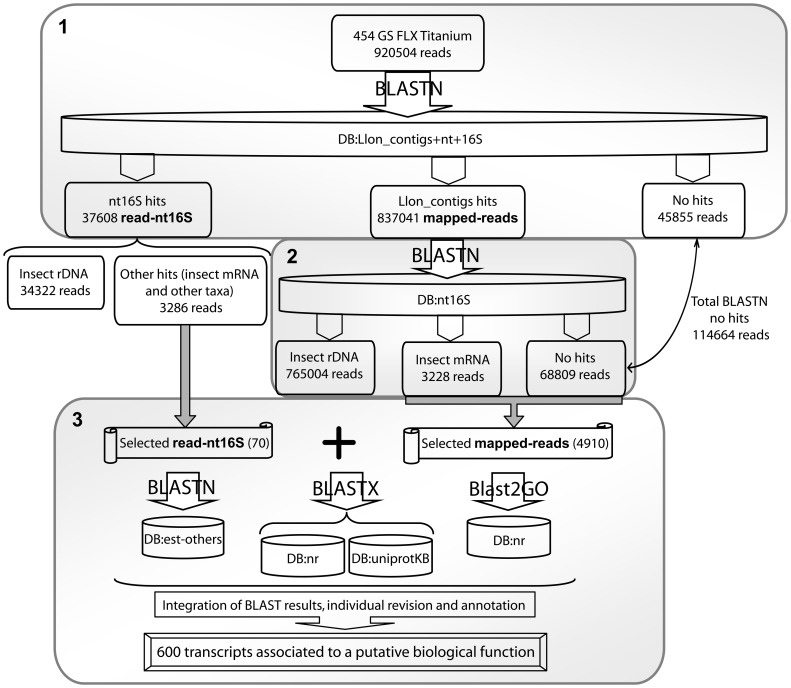
Figure 1. Sequence analysis workflow.
This figure shows an overview of the rationale supporting the analysis of the reads (transcripts) obtained in this study and summarises the different steps that were followed. 1) Indicates the first stage of analysis, in which all the reads were blasted against DB:Llon_contigs+nt+16S. Results were classified in three datasets: reads that mapped to Llon_contigs by sequence homology (mapped-reads); reads that showed homology to nt16S (read-nt16S); and reads that returned no significant hits (no hits) (see text for details). 2) Indicates the second stage of analysis, in which the mapped-reads dataset was blasted against DB:nt16S (see text for details). 3) Indicates the third stage of analysis, in which selected reads (mapped-reads and read-nt16S) were separately blasted against three databases (est-others, nr and uniprotKB) and annotated using Blast2GO (see text for details). The ‘selected mapped-reads (4910)’ excluded mapped-reads 15379, 23694, 25834, 27903, 27904 and 9281, which showed homology to insect rDNA after BLASTN against DB:nt16S, and contig 31202, with unknown function after BLASTN against DB:nt16S (see text for details). The ‘selected read-nt16S (70)’ included read-nt16S that did not show homology to either insect rDNA or taxa other than insects (see text for details).