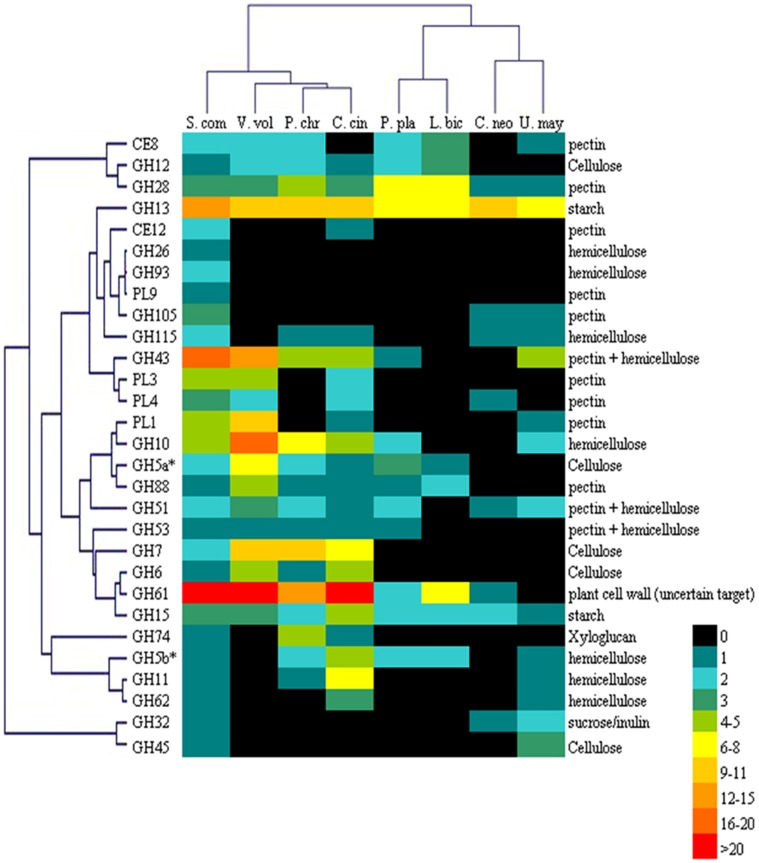
Figure 2. Double clustering of the carbohydrate-cleaving families of 8 basidiomycete genomes.
Top tree: S. com. for Schizophyllum commune; V. vol. for Volvariella volvacea; P. chr. for Phanerochaete chrysosporium; C. cin. for Coprinopsis cinerea; P. pla. for Postia placenta; L. bio. for Laccaria bicolor; C. neo. for Cryptococcus neoformans; U. may. for Ustilago maydis. Left tree: the enzyme families are represented by their class (GH for glycoside hydrolase; PL for polysaccharide lyase; CE for carbohydrate esterase) and family number according to the carbohydrate-active enzyme database [23]. Right side: known substrate of CAZy families. Abundance of the different enzymes within a family is represented by a colour scale from 0 (black) to >20 (red) occurrences per species.