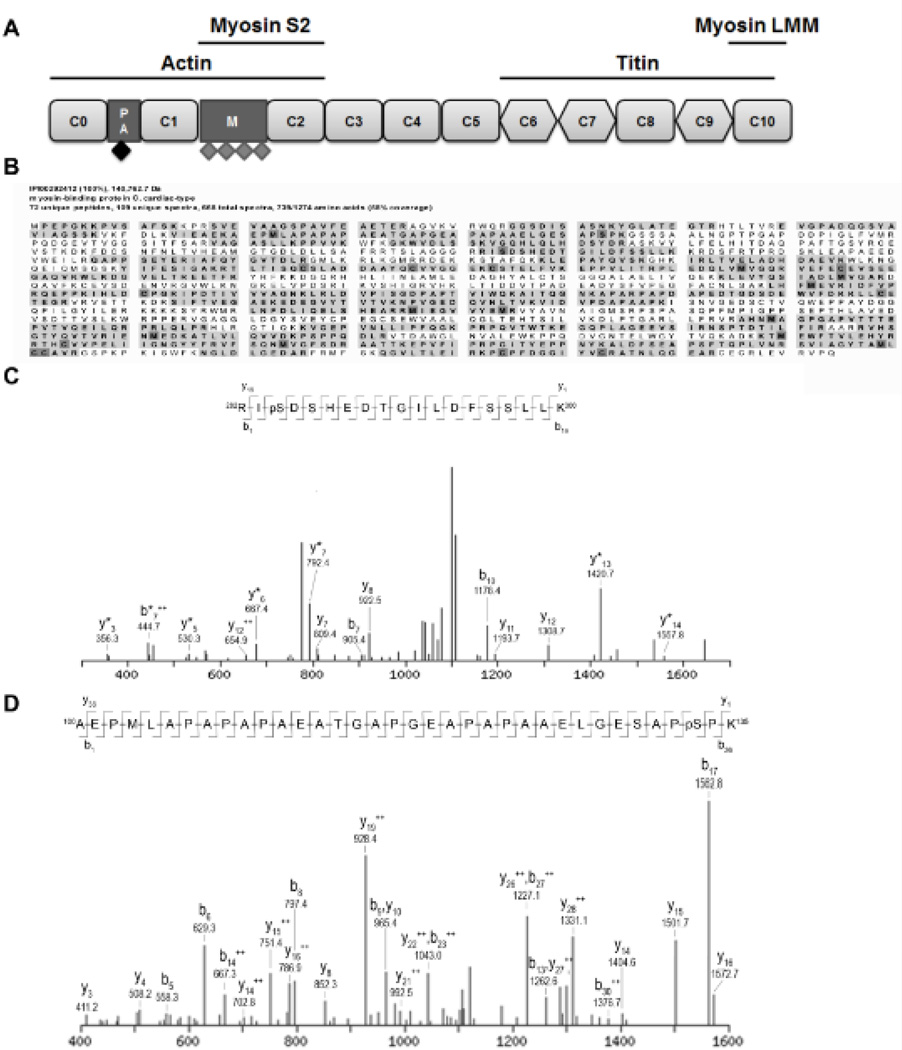
Figure 1. Identified phosphorylated peptides from cMyBP-C.
A. Schematic domain structure of cMyBP-C. Cardiac MyBP-C consists of 8 Ig (rounded rectangle) and 3 fibronectin (diamond) domains labeled C0 (N-terminus) through C10 (C-terminus). Two additional domains are present in the N-terminal part of the protein, the Proline-Alanine rich region (PA) and the M-domain (M). Four previously described phosphorylation sites, Ser275, Ser284, Ser304 and Ser311 (indicated by grey diamonds), are located in the M-domain. Cardiac MyBP-C was semi-purified from donor and failing human myocardium. After trypsin digestion, peptides were scanned by tandem mass spectrometry for phosphorylated peptides. B. Peptide coverage (bold with gray background) of cMyBP-C semi-purified from donor myocardium. 58% of the total amino acid sequence was covered by the peptides identified by the MS/MS analysis. Modified amino acids are indicated in dark gray. C. Fragmentation spectrum with b and y ions indicated of peptide spanning from amino acid 282–300 showed a phosphorylated serine at position 284 in cMyBP-C from donor myocardium. D. Fragmentation spectrum of peptide spanning from amino acids 100–135 revealed a novel phosphorylation site on serine 133 in cMyBP-C semi-purified from donor heart tissue. Serine 133 is located in the PA region (indicated by black diamond in Figure 1A).