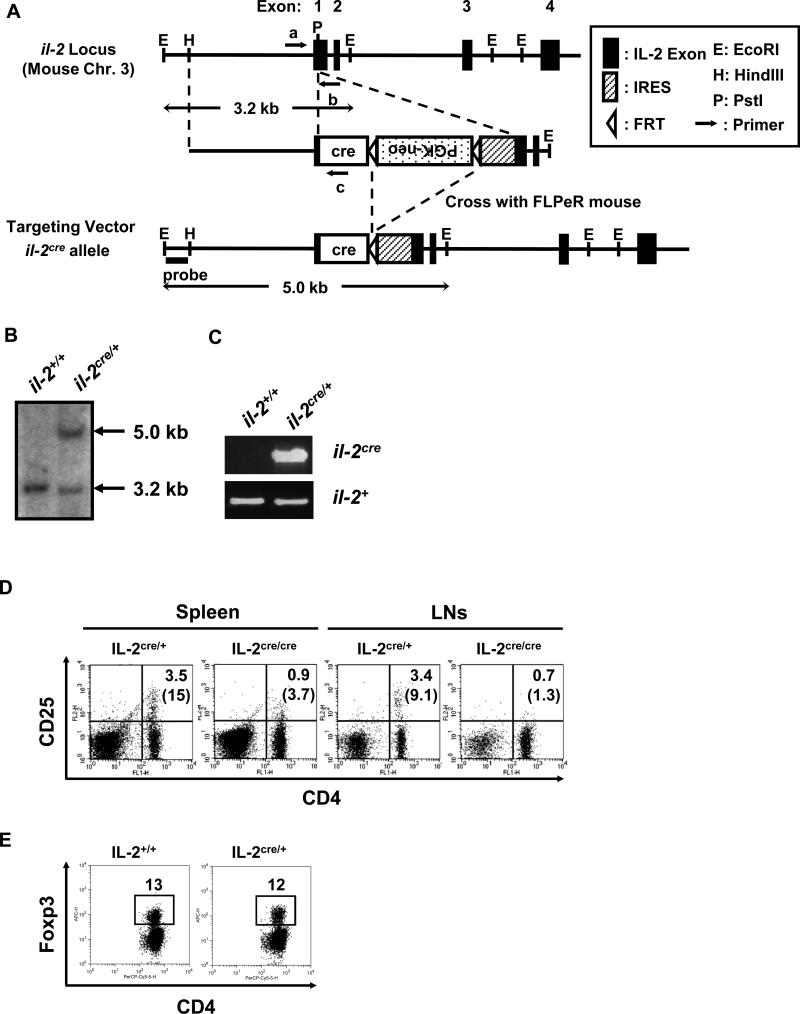
Figure 1. Generation of IL-2cre knock-in mice.
(A) Design of cre knock-in at il-2 locus. Mouse il-2 locus is schematically presented (closed boxes show exons from the wildtype gene) with relevant restriction enzyme sites; the targeting vector and the il-2cre allele (after removal of the FRT- flanked neo gene) are also shown. Arrows represent primers used for genotyping in (C). (B) Southern blotting analysis of tail genomic DNA for il-2cre knock-in. DNA hybridization was performed with the outside probe indicated in (A). The 3.2 kb band corresponds to the DNA fragment derived from wildtype il-2 allele, and the 5.0 kb band corresponds to the il-2cre allele after homologous recombination with the targeting vector. (C) PCR genotyping of tail genomic DNA for IL-2cre mice. Exon 1 with inserted cre (il-2cre) was detected by the primers a and c (shown in Fig. S1A). Exon 1 from wildtype allele (il-2+) was detected by the primers a and b. (D) Spleen and lymph nodes from IL-2cre/+ and litter mate IL-2cre/cre mice were analyzed for the expression of CD4 and CD25. Numbers indicate percentages of CD4+CD25+ cells among total cells. Parenthesized numbers indicate the percentages of CD25+ cells among CD4+ T cells. (E) Expression of Foxp3 by splenic CD4 T cells from wildtype and IL-2cre/+ mice.