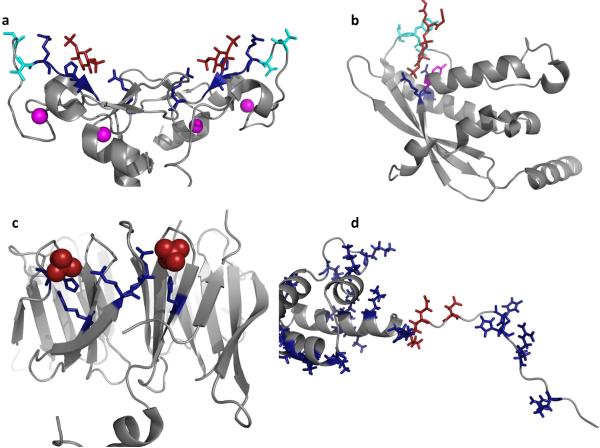
Figure 5. Mechanisms and models of PI(3)P binding by FYVE, PX, PROPPIN, RxLR and WY domains.
PI(3)P binding has been attributed to at least three distinct eukaryotic domains including FYVE, PX, and PROPPINS while pathogens utilize the RxLR or RxLxE/D/Q motif and/or WY domains. (a) The homodimeric structure of the EEA1 FYVE domain is shown (PDB ID: 1JOC), which binds PI(3)P (shown in red) in a sideways fashion where both electrostatic and H-bonding with PI(3)P can be optimized while achieving hydrophobic interactions through the turret loops [39]. Cationic residues that coordinate the PI(3)P headgroup are shown in blue, hydrophobic residues that penetrate into the hydrocarbon portion of the membrane are highlighted in cyan, and the zinc ions coordinated by the FYVE domains for structural stability are shown in magenta. (b) The p40phox PX domain (PDB ID: 1H6H) X-ray structure was solved bound to PI(3)P. Here, two critical arginine residues (shown in blue) mediate the recognition of PI(3)P, while a tyrosine residue (magenta) forms the bottom of the PI(3)P binding pocket. The PI(3)P binding pocket is deeper than that observed for the FYVE domain and harbors two loop regions that have been shown to facilitate nonspecific electrostatic interactions and membrane penetration (hydrophobic residues in cyan) [57]. (c) The yeast PROPPIN structure from Hsv2 (Homologous with SVP1), which is a key autophagic sensor protein, was solved bound to two sulfate ions (PDB ID: 4EXV). PROPPINs are PI(3)P sensors necessary for autophagy and are conserved from yeast to humans. The PROPPIN structure is a β-propeller composed of seven blades where two PI(3)P binding sites (sulfate ions in red and coordinating residues in blue) were discovered on blades five and six. In a similar manner to the FYVE and PX domains, a hydrophobic loop between blades five and six promotes membrane penetration of Hsv2, which is essential to act in concert with PI(3)P binding for membrane recognition and full autophagic function in yeast. (d) Shown is the N-terminal unstructured region and WY domain of Avr1b from P. sojae, which was modeled and generated using the available P. infestans Avr3a structure (PDB ID: 2LC2) [25]. The N-terminus harbors the RxLR motif (Arg residues in blue on the left) as well as number of other cationic (blue) and anionic (red) residues that may be critical tophosphoinositide binding. The WY domain of Avr1b is shown to highlight the cationic nature of this module (Arg, Lys and His shown in blue). In contrast to FYVE, PX, and PROPPINs a distinct phosphoinositide binding pocket or region is not obvious and nonspecific electrostatic association may occur based upon lipid charge.