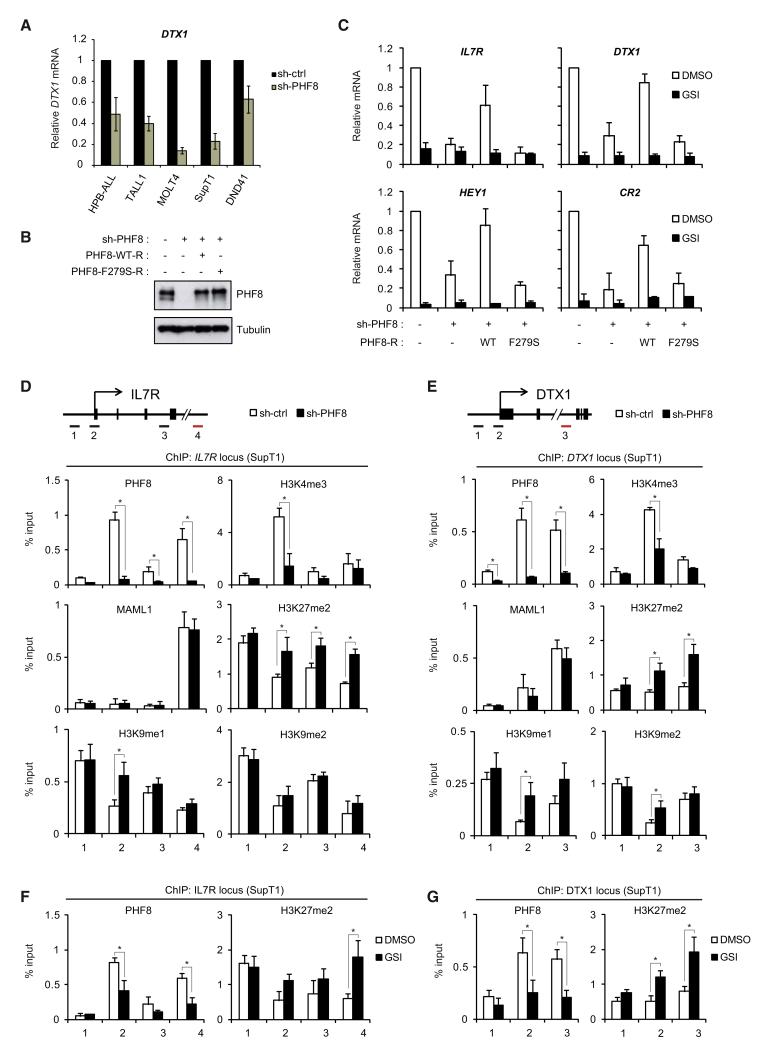
Figure 6. PHF8 Demethylase Activity Is Required for Notch-Mediated Activation of Its Target Genes.
(A) PHF8 is required for Notch-responsive gene expression in T-ALL cell lines. HPB-ALL, TALL1, MOLT4, SupT1 and DND41 were transduced with control or PHF8 shRNA. Expression of DTX1 was measured by quantitative RT-PCR (Q-RT-PCR) and normalized to GAPDH. See also Figure S5A for other Notch-responsive genes.
(B) SupT1 expressing PHF8 shRNA were transduced with shRNA-resistant wide-type (WT) or catalytic inactive (F279S) PHF8 mutant. Endogenous and ectopically expressed PHF8 levels were determined by WB.
(C) mRNA levels of IL7R, DTX1, HEY1, and CR2 were measured by Q-RT-PCR in SupT1 cells coexpressing PHF8 shRNA and the indicated PHF8 construct (from B). Shown are means ± SD (n = 3).
(D and E) ChIP assays for PHF8, MAML1, H3K4me3, H3K27me2, H3K9me1, and H3K9me2 at the IL7R locus (C) and DTX1 locus (D) were performed in SupT1 cells expressing control or PHF8 shRNA (shown as percentage relative to input DNA, n = 3, *p < 0.05). The upper panel depicts the locus structure and positions of the PCR amplicons. See also Figure S5C for H4K20me1 levels. H3K27me2/me3 ChIP results at other Notch-target loci are shown in Figures S5D and S5E.
(F and G) ChIP assays were performed in SupT1 cells treated with DMSO or GSI. Results for PHF8 and H3K27me2 at the IL7R locus (E) and DTX1 locus (F) are shown. ChIP primers described in (D) and (E) were used (n = 3, *p < 0.05). See also Figure S5B for PHF8 effects on a Notch-responsive reporter.