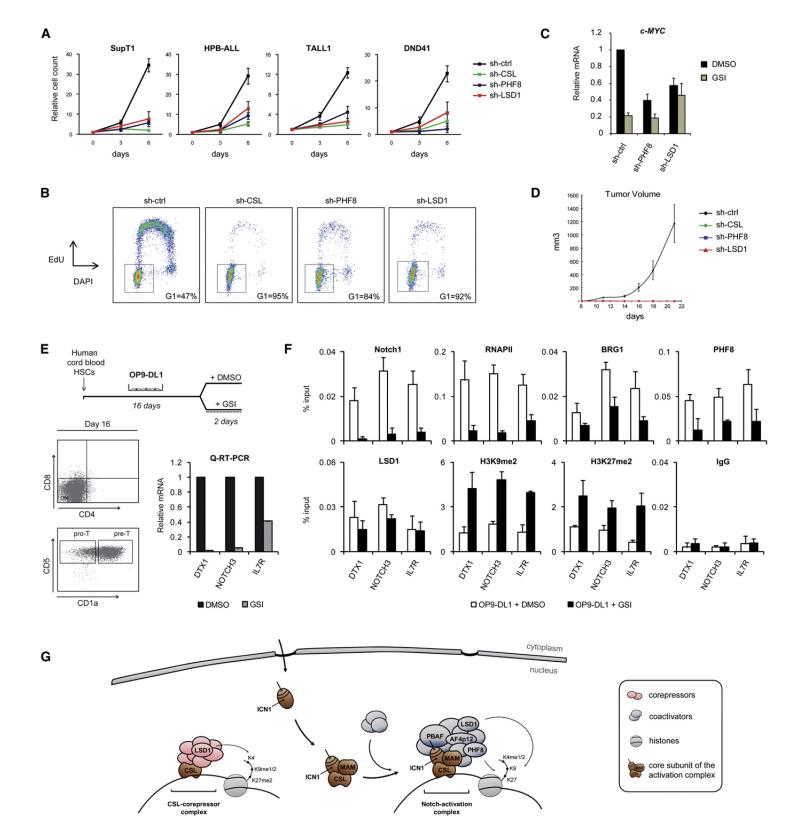
Figure 7. Functional Relevance of Notch Cofactors in T-ALL Proliferation and Gene Expression during T Cell Development.
(A) LSD1 and PHF8 are required for NOTCH1-dependent T-ALL cells growth. SupT1, HPB-ALL, TALL1, and DND41 cells were transduced with control, CSL, PHF8 or LSD1 specific shRNA. One week posttransduction, cell count proliferation assays were performed (n = 3). The observed effects of CSL, PHF8, and LSD1 depletion is significant (p < 0.05).
(B) Effects of PHF8 and LSD1 depletion on cell cyle progression. SupT1 cells expressing control, CSL, PHF8 or LSD1 shRNA were pulse labeled with EdU for 2 hr. DNA synthesis (EdU incorporation, y axis) and total DNA content (DAPI staining, x axis) was measured by flow cytometry. The gate and percentage indicate cells in G0/G1 phase. See also Figure S6A for CSL, LSD1, and PHF8 depletion effects on cell cycle in a panel of NOTCH1-dependent or independent T-ALL cell lines.
(C) Notch-mediated expression of c-MYC in T-ALL cells requires PHF8 and LSD1. Quantitative RT-PCR analysis of c-MYC expression was performed in TALL1 expressing control, PHF8 or LSD1 shRNA and treated with DMSO or GSI (n = 3).
(D) Depletion of PHF8 and LSD1 impairs T-ALL progression in vivo. SupT1 expressing control, CSL, LSD1 or PHF8 shRNA were implanted subcutaneously in SCID mice (n = 5). Xenograft tumor volume was monitored over 21 days. See also Figure S6B.
(E) Hematopoietic stem cells (HSCs) from human cord blood were induced to differentiate to the T cell lineage in OP9-DL1 cocultures. T cell precursors (pre-T) were further treated with DMSO or GSI (upper panel). Cell surface expression of CD4, CD8, CD5, and CD1a at day 16 was analyzed by flow cytometry (Dot plots, lower panel). Expression of DTX1, NOTCH3, and IL7R was analyzed by quantitative RT-PCR on total RNA extracted from DMSO or GSI-treated pre-T (Graphs, lower panel).
(F) Notch cofactors are recruited to target genes during T cell development. ChIP experiments were performed in pre-T from (E) using the indicated antibodies. ICN1-binding region at the DTX1, NOTCH3, and IL7R locus was PCR amplified from the precipitated and input DNA. An irrelevant antibody (IgG) was used as a control. Shown are means ± SD (n = 2).
(G) Model for the regulation of Notch-target genes.