Yeast Mge1, the cochaperone of mitochondrial heat shock protein 70 (mHsp70), is essential for exchanging ATP for ADP on mHsp70 and thus for recycling of mHsp70 for mitochondrial protein import and folding. Mge1 acts as an oxidative sensor to regulate mHsp70 function.
Abstract
Despite the growing evidence of the role of oxidative stress in disease, its molecular mechanism of action remains poorly understood. The yeast Saccharomyces cerevisiae provides a valuable model system in which to elucidate the effects of oxidative stress on mitochondria in higher eukaryotes. Dimeric yeast Mge1, the cochaperone of heat shock protein 70 (Hsp70), is essential for exchanging ATP for ADP on Hsp70 and thus for recycling of Hsp70 for mitochondrial protein import and folding. Here we show an oxidative stress–dependent decrease in Mge1 dimer formation accompanied by a concomitant decrease in Mge1–Hsp70 complex formation in vitro. The Mge1-M155L substitution mutant stabilizes both Mge1 dimer and Mge1–Hsp70 complex formation. Most important, the Mge1-M155L mutant rescues the slow-growth phenomenon associated with the wild-type Mge1 strain in the presence of H2O2 in vivo, stimulation of the ATPase activity of Hsp70, and the protein import defect during oxidative stress in vitro. Furthermore, cross-linking studies reveal that Mge1–Hsp70 complex formation in mitochondria isolated from wild-type Mge1 cells is more susceptible to reactive oxygen species compared with mitochondria from Mge1-M155L cells. This novel oxidative sensor capability of yeast Mge1 might represent an evolutionarily conserved function, given that human recombinant dimeric Mge1 is also sensitive to H2O2.
INTRODUCTION
Mitochondria are essential organelles involved in many cellular processes, such as energy metabolism and apoptosis. Although the mitochondrion has its own genome, it depends on the nucleus for optimal functioning (Chacinska et al., 2009). Based on their signal sequence, mitochondrial proteins encoded by nuclear DNA are targeted to different subcompartments of mitochondria through a translocase system present on outer and inner mitochondrial membranes known as the translocase of outer membrane (TOM) and translocase of inner membrane (TIM) complexes, respectively (Schulke et al., 1997, 1999; Endo et al., 2003; Kutik et al., 2007; Neupert and Herrmann, 2007). Targeting of precursor protein to the matrix involves an interplay among many proteins; however, the final step of this process is mediated by Tim44 and a translocation motor that contains mitochondrial heat shock protein 70 (mHsp70), Pam16, Pam18, and the nucleotide exchange factor Mge1 (Azem et al., 1997; Mokranjac et al., 2007; Stojanovski et al., 2007; Schiller et al., 2008). Hsp70, in combination with Tim44, binds to the emerging end of the transit peptide from the TIM channel in an ATP-dependent manner, and the ATPase cycle of mHsp70 leads to pulling or vectorial translocation of preproteins across the inner mitochondrial membrane (Matouschek et al., 2000; Okamoto et al., 2002; Liu et al., 2003). Mge1, a component of this translocation motor, accelerates the exchange of ATP for ADP on mHsp70 and promotes a change from the high–substrate affinity conformation of mHsp70 to a lower–substrate affinity form with a concomitant release of precursor protein from mHsp70 to begin the next round of translocation (Deloche and Georgopoulos, 1996; Schneider et al., 1996; Miao et al., 1997; Sakuragi et al., 1999).
Mge1 is a homologue of bacterial GrpE and interacts with mHsp70 to facilitate importation across the inner mitochondrial membrane. Yeast Mge1 shares 57% similarity with bacterial GrpE and can functionally substitute the Escherichia coli GrpE, demonstrating the conserved mechanism of action during evolution. Functional Mge1 exists as a dimer, and this dimerization seems to be critical for its interaction with mHsp70, as the monomeric form of Mge1 fails to interact with mHsp70, resulting in a delay in ATPase cycle and chaperonic function of mHsp70 (Moro and Muga, 2006). Besides acting as ATP–ADP exchanger, Mge1 acts as a thermosensor in bacteria and yeast (Grimshaw et al., 2001, 2003; Groemping and Reinstein, 2001). At high temperatures, Mge1 exists as a monomer and fails to interact with mHsp70 (Grimshaw et al., 2001; Moro and Muga, 2006). Further, temperature-sensitive mutants of Mge1 accumulate precursors in the cytosol, aggregate proteins in the matrix, and have reduced nucleotide-dependent dissociation of mHsp70 from Tim44 (Laloraya et al., 1995). It is also involved in maintenance of mitochondria during heat stress, sorting of proteins, degradation of misfolded proteins, and the formation of iron–sulfur clusters (Westermann et al., 1995; Dutkiewicz et al., 2006).
Reactive oxygen species (ROS) have been implicated in aging, as well as in a host of diseases (Mammucari and Rizzuto, 2010). The electron transport chain present in mitochondria is the major source for the generation of reactive oxygen species, and there has been an increased focus on identifying the protein(s) that alter activity during oxidative stress, in order to understand mitochondria-mediated ROS signaling and diseases (Drechsel and Patel, 2009; Murphy, 2009). Decreased import of mitochondrial DNA repair enzyme in aged samples (Szczesny et al., 2003) and reduced import of preproteins under pro-oxidant conditions directly imply a role for ROS in mitochondrial protein import (Wright et al., 2001). Further, oxidative stress impairs the growth of yeast (Demasi et al., 2006). However, no protein import factor or receptor of mitochondria has been implicated in the oxidative stress response in either lower or higher eukaryotes.
In this study, we find that recombinant yeast and human Mge1 fail to dimerize and interact with mHsp70 in the presence H2O2 in vitro. In addition, by using Saccharomyces cerevisiae as a model system, we show that oxidative stress reduces interaction of mHsp70 with Mge1 in vivo. Of interest, a single amino acid substitution from methionine to leucine in Mge1 renders this protein resistant to oxidative stress in vitro and in vivo. Our studies suggest that Mge1 acts as a novel oxidative sensor of mitochondria to regulate mitochondrial protein import and folding.
RESULTS
Oxidative stress reduces Mge1 and mHsp70 complex formation in vitro
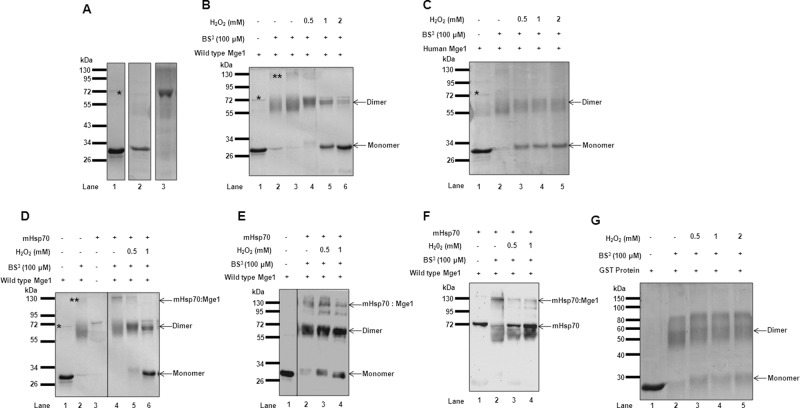
Some proteins undergo oxidation when they are exposed to H2O2 in vitro (Men and Wang, 2007). To investigate whether Mge1 responds to oxidative stress, we treated purified recombinant Mge1 (Figure 1A) with increasing concentrations of H2O2, followed by cross-linking with the amine reactive cross-linker bis(sulfosuccinimidyl) suberate (BS3). The samples were resolved on SDS–PAGE, and Coomassie-stained gels were analyzed (Figure 1B). Mge1 migrates as a 27-kDa monomer in the absence of a cross-linker (Figure 1, A and B). In the presence of BS3, Mge1 migrates as a ∼60-kDa homodimer, reflecting the efficiency of the cross-linking agent. However, in the presence of H2O2, the appearance of monomer increased in a concentration-dependent manner despite the presence of cross-linker (Figure 1B, compare lane 3 with lanes 4–6). To determine whether the Mge1 response to H2O2 is conserved across species, we repeated the experiment, using purified human Mge1 (Figure 1A). Consistent with the observations made using yeast Mge1, the dimer form of human Mge1 is also susceptible to H2O2 (Figure 1C). These results suggest that Mge1 loses its dimer formation ability in the presence of oxidative stress triggered by H2O2.
FIGURE 1:
H2O2 reduces Mge1 dimer formation and Mge1–mHsp70 complex formation. (A) Purified recombinant proteins of yeast Mge1 (lane1), human Mge1 (lane 2), and yeast mHsp70 (lane 3) were resolved on SDS–PAGE and Coomassie stained. (B) Purified recombinant yeast Mge1 (lane 1) was incubated in the presence (lanes 4–6) and absence (lanes 2 and 3) of H2O2 for 20 min at room temperature in 20 mM sodium phosphate buffer (pH 7.4) and cross-linked with 100 μM BS3 (lanes 2–6) for 20 min. The cross-linking reactions were quenched, and adducts were resolved on SDS–PAGE and Coomassie stained. (C) Coomassie-stained, cross-linked adducts of human Mge1 in the presence (lanes 3–5) or absence of (lane 2) of H2O2. (D) Purified Mge1 was treated with (lanes 5 and 6) or without (lanes 1–4) increasing concentrations of H2O2 (0.5 and 1 mM) for 20 min at room temperature. The samples were incubated with purified mHsp70 (lanes 3–6) for 10 min, followed by cross-linking with BS3 (lanes 2 and 4–6) for 20 min at room temperature and were quenched with Tris-HCl buffer. The cross-linked adducts were separated on SDS–PAGE and Coomassie stained or Western transferred and probed with antibodies specific for Mge1 (E) or mHsp70 antibodies (F). (G) Purified recombinant GST protein was subjected to increasing concentrations of H2O2 and cross-linked, and cross-linked adducts were separated and stained with Coomassie. Asterisk, copurified DnaK with recombinant yeast Mge1; double asterisks, the DnaK–Mge1 complex.
Because the dimerization of Mge1 was shown to be crucial for its interaction with mHsp70, we evaluated the effect of H2O2 on this interaction, using recombinant purified yeast Mge1 and mHsp70 (Figure 1A). We incubated recombinant mHsp70 with Mge1 that was either left untreated or treated with H2O2 (Figure 1D). The reaction mixtures were cross-linked, resolved on SDS–PAGE, and stained with Coomassie blue (Figure 1D). A high–molecular weight cross-linked band was observed at ∼130 kDa upon incubation of Mge1 with mHsp70 in the absence of H2O2 (Figure 1D, lane 4). Of interest, formation of this complex decreases with prior treatment of Mge1 with increasing concentration of H2O2 (Figure 1D, compare lane 4 with lanes 5 and 6). To confirm the constituents of the 130-kDa complex, we carried out Western blotting using antibodies specific for Mge1 (Figure 1E) and mHsp70 (Figure 1F). Both antibodies detected a similar strong, high–molecular weight (130 kDa) band in the absence of H2O2, suggesting that indeed Mge1 and mHsp70 exist in a high–molecular weight complex. To rule out the possible interference of H2O2 with BS3 cross-linking efficiency, we used glutathione S-transferase (GST), a dimeric protein (Fancy and Kodadek, 1999), as a control and cross-linked with BS3 in the presence and absence of H2O2. Most of the GST remains as a dimer in the presence of H2O2, indicating that BS3 is not affected by H2O2 (Figure 1G). Taken together, our results clearly show that Mge1 is able to modulate its interaction with mHsp70 in a manner that is dependent on the oxidative stress in vitro. Mge1 is able to switch from an active dimer to an inactive monomer form by sensing the oxidative stress. Of interest, we find that recombinant Mge1 copurifies with a small amount of bacterial DnaK protein, the bacterial homologue of eukaryotic mHsp70 (Figure 1, asterisk). DnaK also forms a high–molecular weight complex with Mge1 (Figure 1B, double asterisks), and we find that this interaction is dependent on the Mge1 dimer, as pretreatment of Mge1 with H2O2 decreases the amount of the Mge1–DnaK complex (Figure 1B), thus indicating that the mechanism might have been conserved across evolution.
Methionine 155 in Mge1 is prone to oxidative modification in vitro
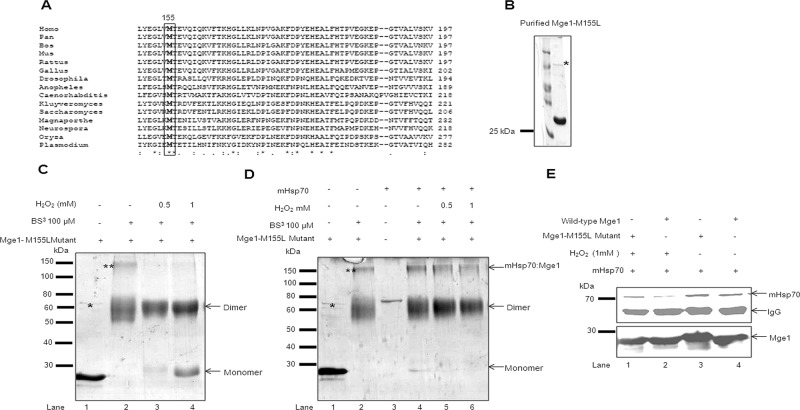
Because pretreatment of Mge1 with H2O2 induces conformational change and affects its interaction with mHsp70, we further investigated the amino acid residues in Mge1 that are prone to oxidative modification. Generally, cysteine and methionine are most susceptible to oxidation, followed by tryptophan, histidine, and tyrosine (Kim et al., 2001). Surprisingly, yeast Mge1 protein is devoid of cysteine and contains only one methionine at the 155 position, and this methionine residue is highly conserved across species (Figure 2A). To test whether methionine 155 undergoes oxidative modification upon exposure to H2O2, we mutated methionine 155 to leucine (Mge1-M155L mutant) and investigated its response to oxidative stress in vitro. Recombinant purified Mge1-M155L mutant (Figure 2B) was treated with increasing concentrations of H2O2 and incubated without (Figure 2C) or with mHsp70 (Figure 2D). The cross-linked samples were resolved on SDS–PAGE and Coomassie stained for further analysis. Mge1-M155L mutant also forms an ∼60-kDa dimer upon cross-linking (Figure 2C, lane 2), but we observed low amounts of dimer dissociation in the presence of H2O2 (Figure 2C, lanes 3 and 4). In addition, this mutant forms a relatively stable complex with mHsp70 when compared with wild type even after pretreatment with H2O2 (Figures 2D, lanes 4–6, and 1D, lanes 4–6). We obtained similar results when we performed Western immunoprecipitation analysis of H2O2-treated, purified wild-type and Mge1-M155L mutant in the presence of mHsp70. We observed reduced coprecipitation of mHsp70 with Mge1 antibody when we treated wild-type Mge1 with H2O2 as compared with untreated or Mge1-M155L treated with H2O2 (Figure 2E). In addition, we modeled the structure of yeast Mge1 using the I-TASSER (Zhang, 2008; Roy et al., 2010) server and found that the methionine 155 position is present in the helical region of the protein (Supplemental Figure S1). The helical region is downstream of the dimerization domain. Of interest, this residue is present on the outer surface of the helical region and may be available for possible modifications. Like wild-type Mge1, Mge1-M155L also copurifies with a small amount of DnaK and forms a high–molecular weight, 130-kDa complex upon cross-linking (indicated by double asterisks). However, this complex is more susceptible to H2O2 treatment when compared with the Mge1-M155–mHsp70 complex (Figures 1C, lanes 2–4, and 2D, lanes 4–6). These results suggest the likely involvement of methionine 155 in the oxidative response of Mge1.
FIGURE 2:
Mge1-M155L mutant is resistant to H2O2-induced oxidative stress. (A) A multiple sequence alignment of C-terminal of Mge1 (149–197) across eukaryotic species. Black-shaded box represents the conserved methionine amino acid. (B) Purified recombinant Mge1 (M155L-Mge1) was resolved on SDS–PAGE and Coomassie stained. (C) Purified Mge1-M155L was preincubated in 20 mM sodium phosphate (pH 7.4) buffer in the presence (lanes 3 and 4) or absence (lanes 1 and 2) of H2O2 for 20 min, followed by cross-linking with BS3. (D) Purified Mge1-M155L was preincubated in 20 mM sodium phosphate (pH 7.4) buffer in the presence (lanes 3–5) or absence (lanes 1) of H2O2 for 20 min before incubation with mHsp70 (lanes 2–5) for 10 min. Then the samples were cross-linked with BS3 final concentration at 100 μM. Samples were incubated at room temperature for 30 min and quenched by Tris-HCl buffer. The samples were resolved on SDS–PAGE and Coomassie stained. (E) Purified wild type (lanes 2 and 4) and Mge1-M155L (lanes 1 and 3) were treated with 1 mM H2O2 for 20 min, followed by incubation with mHsp70 for 10 min. Then the samples were immunoprecipitated with anti Mge1 antibodies and probed with antibodies specific for mHsp70 (top) and Mge1 (bottom). This blot is representative of two independent experiments. Asterisk, the copurified DnaK with recombinant yeast Mge1-M155L; double asterisks, the DnaK–Mge1-M155L complex.
Altered structure of Mge1 with H2O2 treatment
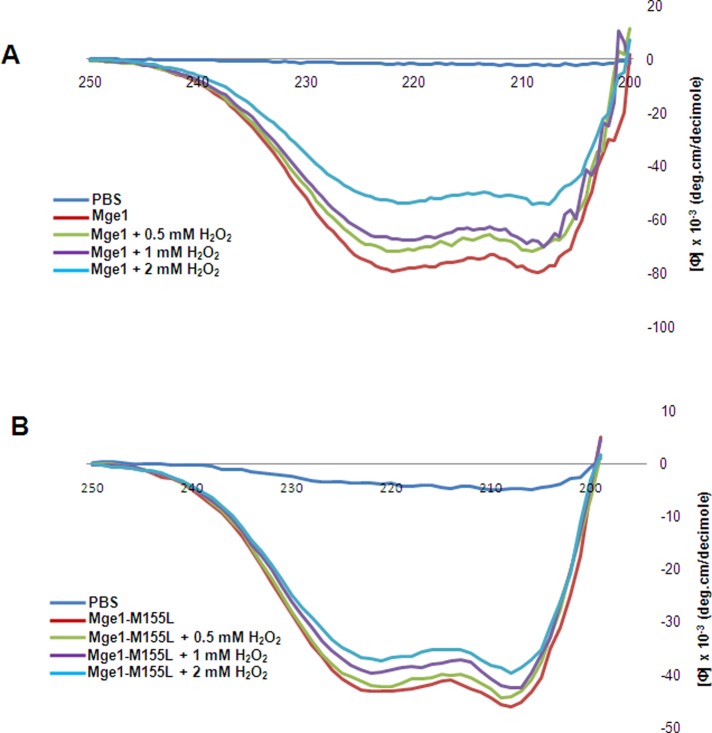
Because H2O2 induced the structural transition of Mge1 from dimer to monomer, we used circular dichroism (CD) spectroscopy to analyze structural changes in Mge1 in the presence of H2O2 (Figure 3) and quantified secondary structural alterations using CDNN 2.1 software. The CD spectra of wild-type Mge1 exhibited two negative bands in the ultraviolet region, at 208 and 218 nm. There is a gradual and significant alteration of the spectrum in wild-type Mge1 treated with increasing concentration of H2O2 (Figure 3A). Overall, α-helical, β, and antiparallel turns in wild-type Mge1 are considerably altered with increasing concentration of H2O2 (Table 1). It is evident from the spectrum that the isodichroic point is at 203 nm and is not altered with varying concentration of H2O2, indicating the presence of a two-state conformation. This also suggests that loss of helical stability is possibly due to partial unfolding of Mge1 protein. In the case of Mge1-M155L mutant, the shape of the spectrum is similar to that of the wild type in the absence of H2O2 (Figure 3B). However, treatment of Mge1-M155L mutant with increasing concentrations of H2O2 does not appreciably alter the α-helical and β-sheet structures (Table 2). These results show that M155L mutation stabilizes the Mge1 structure and protects it from H2O2-induced oxidative stress. This is consistent with our earlier observation of a stable Mge1-M155L–mHsp70 complex in the presence of H2O2 in vitro.
FIGURE 3:
Circular dichroism analysis of Mge1 under oxidative stress. Purified and dialyzed Mge1 (40 μM) wild-type and Mge1-M155L mutant samples in phosphate buffer were used for CD spectra analysis at 20°C at a scan rate of 60°C/h in a 2-mm–path-length cuvette. Wild type (A) and mutant Mge1 (B) were pretreated with 0.5, 1, or 2 mM H2O2 for 15 min before spectral analysis.
TABLE 1:
Secondary structural analysis of H2O2-treated recombinant wild-type Mge1 from Figure 3A.
| Wild-type Mge1 | 0.5 mM H2O2 | 1 mM H2O2 | 2 mM H2O2 | |
|---|---|---|---|---|
| Helix (%) | 75.00 | 65.90 | 63.50 | 44.50 |
| Antiparallel (%) | 2.00 | 3.20 | 3.50 | 6.00 |
| Parallel (%) | 2.00 | 3.20 | 3.60 | 6.60 |
| β-Turn (%) | 10.00 | 12.20 | 12.60 | 16.20 |
| Random coil (%) | 11.00 | 15.50 | 16.70 | 26.80 |
| Sum (%) | 100.00 | 100.00 | 100.00 | 100.00 |
All at 200–260 nm.
TABLE 2:
Secondary structural analysis of H2O2-treated recombinant Mge1-M155L mutant from Figure 3B.
| Mge1-M155L | 0.5 mM H2O2 | 1 mM H2O2 | 2 mM H2O2 | |
|---|---|---|---|---|
| Helix (%) | 72.00 | 70.20 | 67.50 | 64.40 |
| Antiparallel (%) | 2.50 | 2.70 | 3.00 | 3.50 |
| Parallel (%) | 2.40 | 2.60 | 3.00 | 3.50 |
| β-Turn (%) | 11.10 | 11.50 | 12.00 | 12.50 |
| Random coil (%) | 12.00 | 13.00 | 14.50 | 16.10 |
| Sum (%) | 100.00 | 100.00 | 100.00 | 100.00 |
All at 200–260 nm. The data are analyzed by Web-based software CDNN 2.1.
Mge1 acts as an oxidative sensor in vivo
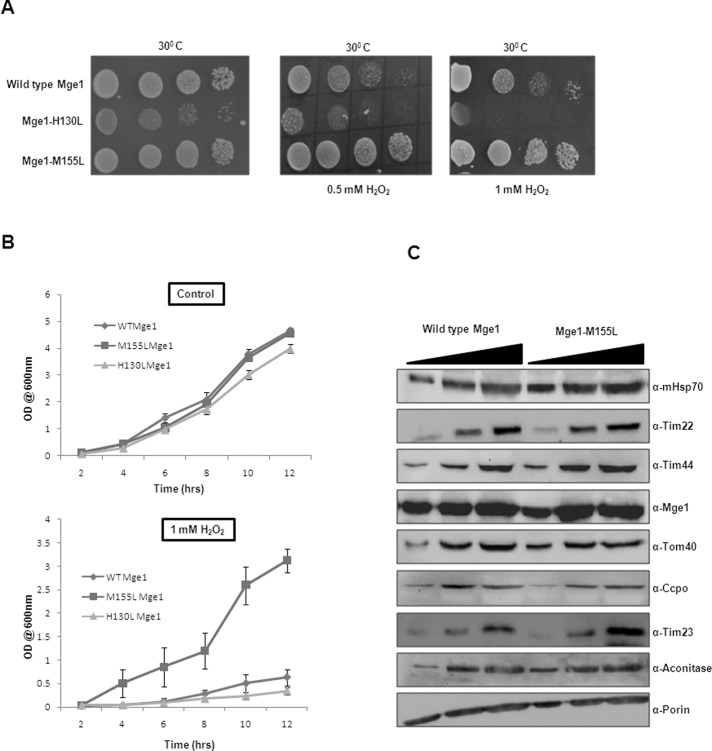
To understand the in vivo significance of Mge1 in oxidative stress, we generated chromosomal MGE1–deleted haploid yeast strains harboring and expressing MGE1 wild-type or MGE1 mutants under a high-copy plasmid. Methionine, cysteine, tryptophan, and histidine are amino acids that can be modified due to oxidative stress. Mge1 does not have any tryptophan or cysteine but has one methionine and four histidines. Because histidine is one of the other possible amino acid that can be modified by oxidative stress, we mutated histidine at the 130 position with leucine (Mge1-H130L) and used this as a control for any nonspecific activity of the Mge1-M155L mutant. We compared the phenotypes of wild-type MGE1 strain with those of MGE1 mutants on Synthetic Complete (SC)-Leu plates supplemented with 0.5 or 1.0 mM H2O2 (Figure 4A). Exponentially growing yeast cells expressing either wild-type Mge1 or Mge1 mutants were serially diluted and spotted on SC-Leu plates with increasing concentration of H2O2. We found that wild-type Mge1 and Mge1-M155L mutant have comparable growth on SC-Leu. However, Mge1-H130L mutant has poor growth. Wild-type Mge1 is sensitive to H2O2, as its growth is retarded even at 0.5 mM H2O2. Most important, Mge1-M155L mutant has considerable growth in the presence of both 0.5 and 1 mM H2O2 when compared with wild type. The slow growth of Mge1-H130L mutant is further exacerbated in the presence of H2O2. To confirm the results obtained on plates, we grew the foregoing strains in liquid SC-Leu medium with or without 1 mM H2O2. All of the strains display similar growth profiles in SC-Leu medium (Figure 4B). However, consistent with our previous results on SC-Leu plates with H2O2, Mge1-M155L mutant is relatively more resistant to H2O2, whereas both wild-type and Mge1-H130L mutant are inhibited by H2O2. A single point mutation in Mge1 protein at M155L is able to rescue poor growth of wild-type yeast in the presence of H2O2. These results further show that Mge1 plays a significant role in the oxidative stress response and that methionine 155 is crucial for this activity.
FIGURE 4:
Sensitivity of wild-type and mutant Mge1-M155L strains to H2O2. (A) Yeast strains yNB71, yNB72, and yNB73 carrying wild-type MGE1 and MGE1-H130L and MGE1-M155L, respectively, were grown overnight in Sc-Leu medium. Then one OD of cells was serially diluted by 10-fold, and 10 μl of each suspension was spotted on either SD plates or SD plates containing 0.5 or 1 mM H2O2 concentration as indicated in the figure and incubated at 30°C for 3 d. (B) Overnight-grown wild-type or mutant cell lines were freshly diluted into SC-Leu medium in the presence and absence of H2O2 (1 mM) at 30°C. The growth was monitored every 2 h by taking OD600. To derive statistical significance, three independent cultures of wild type and mutant were grown in the presence and absence of H2O2. (C) The indicated amounts of mitochondria isolated from either wild-type Mge1 or Mge1-M155L mutant were analyzed by SDS–PAGE and immunostaining with antibodies against specific mitochondrial proteins.
To exclude the possibility that altered protein expression in Mge1-M155L mutant is responsible for the resistance to oxidative stress, we evaluated the steady-state levels of mitochondrial proteins in wild type and Mge1-M155L mutant by using antibodies specific for porin, Tim44, mHsp70, Tim22, CCPO, aconitase, Tim23, and Tom40 (Figure 4C). As shown in Figure 4C, there is no change in steady-state levels of mitochondrial proteins in Mge1-M155L mutant when compared with wild type.
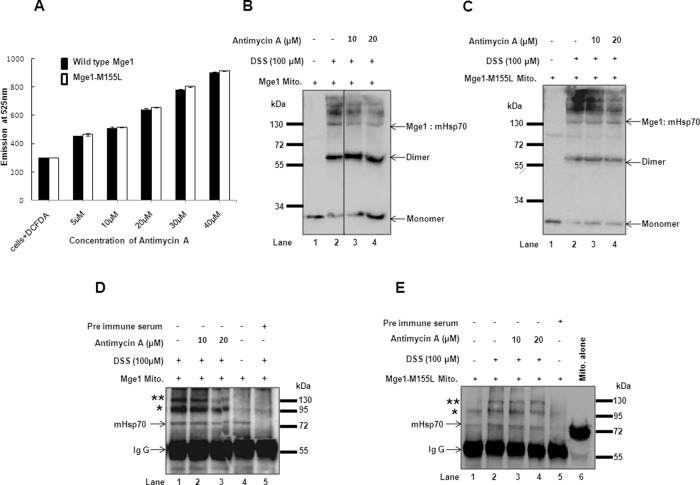
To address whether the resistance of Mge1-M155L mutant to H2O2 is due to stable interaction of mHsp70-Mge1 complex under oxidative stress in vivo, we used antimycin A, an electron transport chain complex III inhibitor that is known to generate ROS in yeast (Smith et al., 2007). Generation of ROS by antimycin A was measured as described in Materials and Methods. Antimycin A induces a similar amount of ROS in all yeast strains tested (Figure 5A). To study the in vivo response of Mge1 to ROS, we isolated mitochondria from wild type and Mge1-M155L mutant and incubated them with increasing concentrations of antimycin A followed by cross-linking with DSS, a membrane-permeable cross-linker (Pierce, Indianapolis, IN; Curran et al., 2002; Mokranjac et al., 2003). We resolved the cross-linked products on SDS–PAGE and Western blotted and probed them with Mge1 antibody in order to study the Mge1 dimer and other interacting protein complexes in the presence and absence of antimycin A (Figure 5, B and C). In the absence of antimycin A but in the presence of cross-linker, three high–molecular weight complexes and one ∼130-kDa complex containing Mge1 were present in both wild type and Mge1-M155L mutant (Figure 5, B and C, lane 2). The 60-kDa complex is likely to be Mge1 dimer, and the high–molecular weight complexes are likely to contain mHsp70 and Mge1 (Moro and Muga, 2006) or Mge1-interacting complexes. However, the formation of high–molecular weight complexes is moderately reduced with antimycin A treatment in wild-type but not in Mge1-M155L mutant mitochondria (Figure 5, B and C, compare lanes 2 and 3 with lane 4). Correspondingly, there is an increase in Mge1 monomer and dimer formation in wild-type but not in mutant mitochondria (Figure 5, B and C, lanes 3 and 4).
FIGURE 5:
Effect of oxidative stress on the oligomeric state and interaction of Mge1 with mHsp70 in mitochondria. (A) ROS measurement was carried out as described in Materials and Methods; the fluorescence signal derived from wild type and Mge1-M55L mutant in the absence or presence of increasing concentrations of antimycin A is shown. Mitochondria were isolated from wild-type (B, D) or Mge1-M155L mutant (C, E) strain. (B, C) Mitochondria were treated without (lanes 1 and 2) or with antimycin A (lanes 3 and 4) and subjected to cross-linking with membrane-permeable cross-linker DSS (lanes 2–4). Samples were incubated at room temperature for 30 min, and the reaction was quenched by addition of Tris-HCl (pH 7.5) buffer. Samples were separated on SDS–PAGE, Western transferred, and probed with antibodies specific for Mge1. (D, E) Wild-type Mge1 (D) and Mge1-M155L mutant (E) mitochondria were treated with or without antimycin A and subjected to cross-linking (D, lanes 1–3 and 5; E, lanes 2–4) with the membrane-permeable cross-linker DSS. Samples were incubated at room temperature for 30 min, and the reaction was quenched by addition of Tris-HCl (pH 7.5) buffer. The samples were subjected to immunoprecipitation with Mge1 antibodies and immunodecorated with mHsp70 antibodies. Preimmune serum was used as a control (lane 5). We loaded 20 μg of mitochondria (E, lane 6) to identify the mobility of mHsp70.
To determine whether Mge1 containing high–molecular weight complexes indeed contain mHsp70, we treated wild-type and mutant mitochondria with antimycin A (10 and 20 μM) and cross-linked them with DSS. Later we solubilized mitochondrial samples, immunoprecipitated them with antibodies specific for Mge1 (Figure 5, D and E), and probed them with mHsp70 antibodies. We were able to detect mHsp70 (Figure 5D, lane 4) in Mge1-mediated immunoprecipitated samples of wild-type mitochondria in the absence of cross-linker. In the presence of cross-linker, we observed mHsp70 in a high–molecular weight complex (Figure 5D, double asterisks). The amount of Mge1–mHsp70 complex (indicated by double asterisks) immunoprecipitated with Mge1 antibodies is consistently reduced by 50–60% in the presence of 20 μM of antimycin A in wild-type mitochondria despite an equal amount of immunoglobulin G in all fractions (Figure 5D). However, we cannot rule out the possibility that one of the complexes (indicates by an asterisk) could be a nonspecific band or Mge1–Hsp70 complex, since background levels of the band are present when the mitochondria sample was used for immunoprecipitation either without cross-linker (Figure 5E, lane 1) or with preimmune serum (Figure 5E, lane 5). Nevertheless, there is no substantial decrease in Mge1–mHsp70 complexes in Mge1-M155L mutant mitochondria samples compared with wild type (Figure 5E). Our observations are consistent with our in vitro findings that Mge1 interaction with mHsp70 diminishes during oxidative stress and that M155L within Mge1 is important for imparting this sensitivity.
Mge1-M155L mutant rescues protein import defect and ATPase activity of Hsp70 during oxidative stress
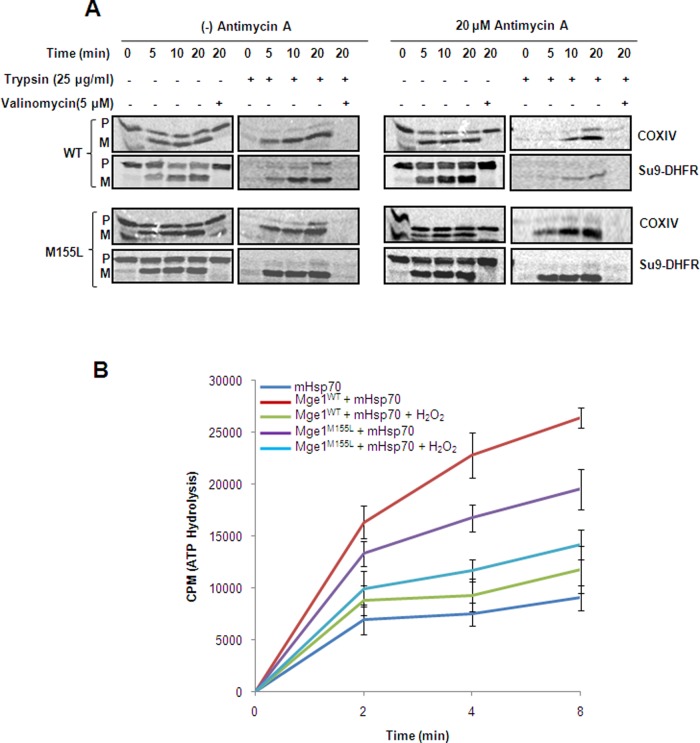
On the basis of our observations so far, we would expect the dissociation of Mge1 from Hsp70 in the presence of oxidative stress to affect protein import. We used the well-established in vitro import assay to determine the effect of antimycin A on protein import. We used [35S]methionine-labeled matrix targeting cytochrome oxidase subunit IV (CoxIV) and Su9-DHFR for import assays. The import of CoxIV and Su9-DHFR is monitored by the processing of N-terminal mitochondrial presequence by mitochondrial processing peptidase and the protection of the mature form from an externally added protease (Sepuri et al., 1998a). We performed kinetics of import of CoxIV and Su9-DHFR in mitochondrial fractions isolated from wild type and Mge1-M155L mutant. As shown in Figure 6A, CoxIV and Su9-DHFR are effectively processed, and the mature form is protected from the externally added protease in wild-type and Mge1-M155L mutant mitochondria (Figure 6A, left). However, imported CoxIV and Su9-DHFR are poorly protected from protease treatment at early time points in wild-type mitochondria pretreated with 20 μM antimycin A (Figure 6A, top right). However, CoxIV and Su9-DHFR are protected at early time points in antimycin A–treated Mge1-M155L mutant sample and are comparable to what is observed in untreated samples at all time points (Figure 6A, bottom). Of interest, the processing of CoxIV and Su9-DHFR does not decrease significantly with antimycin A treatment (Figure 6A, left) in both wild type and Mge1-M155L mutant, indicating that the precursor is not completely translocated into mitochondria in antimycin A–treated wild-type mitochondria (Ungermann et al., 1994). The effect observed with antimycin A is not due to alteration in the membrane potential, as we do not observe any changes in membrane potential (unpublished data). Further, the processing of precursor protein requires membrane potential, as the addition of valinomycin inhibits the processing of preprotein and protection from protease treatment in wild-type and mutant mitochondria (Figure 6A). We conclude that ROS decreases the mHsp70 import cycle and Mge1-M155L mutation suppresses the defect caused by the alteration of the Hsp70–Mge1 interaction.
FIGURE 6:
Effect of oxidative stress on protein import and ATPase activity of mHsp70. (A) Mitochondria isolated from wild type or mutants were either pretreated with 20 μM antimycin A for 20 min or left on ice before import. Mitochondria were treated with valinomycin (5 μm) for 10 min on ice. Reticulocyte lysate–synthesized, 35S-labeled mitochondrial precursor proteins (CoxIV and Su9-DHFR) were imported with or without antimycin A into pretreated wild-type Mge1 and Mge1-M155L mutant mitochondria for different time points. The samples were incubated with (right) and without (left) 25 μg/ml trypsin and analyzed by SDS–PAGE, followed by digital autoradiography as mentioned in Materials and Methods. This is a representative figure of two independent experiments. (B) Purified Mge1 and Mge1-M155L mutant were treated with or without 1 mM H2O2 for 20 min in 20 mM HEPES/KOH, pH 7.2, buffer. Then the proteins were incubated with recombinant Hsp70 in ATPase assay buffer, and we analyzed the release of labeled Pi at different time points as indicated in Materials and Methods.
To further analyze the role of oxidative stress on interaction of Mge1 with mHsp70 function, we used the steady-state ATPase activity of Hsp70 in the presence of Mge1. Mge1 assists in release of ADP for ATP on Hsp70 and thereby accelerates ATP hydrolysis. The ATPase activity of mHsp70 was measured in vitro as described in Materials and Methods. Recombinant mHsp70 alone shows a low level of ATPase activity (Figure 6B). Addition of wild type or Mge1-M155L mutant stimulates the ATPase activity of mHsp70. However, treatment of wild type with H2O2 before incubation with mHsp70 reduces the ATPase activity of mHsp70 significantly when compared with Mge1-M155L mutant. Our results suggest that oxidative stress decreases the interaction of Mge1 with mHsp70 and thereby modulates its function.
DISCUSSION
The mitochondrial electron transport chain is the major source of ROS generation within a cell (Brand, 2010). High levels of ROS lead to alteration in the structure of proteins, lipids, and DNA (Wright et al., 2001; Mammucari and Rizzuto, 2010). Further, oxidative stress in the form of H2O2 reduces the activity of several mitochondrial proteins, such as aconitase, α-ketoglutarate dehydrogenase, and succinate dehydrogenase, in mammalian cells (Bulteau et al., 2003). In addition, accumulation of oxidized proteins in the mitochondria is correlated with aging (Schoneich, 1999). Several lines of evidence suggest that the role of mitochondria-mediated oxidative stress in the progression of several neurological disorders (Lin and Beal, 2006; Zuin et al., 2008). However, very little is known about the role of mitochondria-generated ROS in regulating mitochondrial metabolism and biogenesis. One of the key regulators in mitochondrial biogenesis is translocation of nuclear-encoded proteins into various compartments of mitochondria. It has been shown that redox conditions influence the import of preproteins in higher eukaryotes. Pretreatment of mammalian mitochondria with paraquat, an oxidizing agent, causes the production of ROS at the level of complex I (Cocheme and Murphy, 2008) and causes reduction in the import of pre–ornithine transcarbamylase in intact cells, as well as in vitro. Oxidative stress also leads to a decrease in the processing of preprotein superoxide dismutase in Sf9 cells (Wright et al., 1997; Wright and Reichenbecher, 1999). The mechanism behind the reduced protein import into mitochondria in mammalian cells and in Sf9 cells is not known.
The present study stemmed from the observation that Mge1 acts as a thermosensor during heat stress and undergoes conformational changes that result in reduced ATPase cycle of mHsp70. On the basis of the observation that pro-oxidant conditions also reduce the import of preproteins, we evaluated whether Mge1 has any relevance in protein import during oxidative stress. It is probable that alteration in Mge1 structure may be one of the reasons for alteration in the import of preproteins during oxidative conditions. We showed for the first time that Mge1, a cochaperone of mHsp70, can act as an oxidative sensor both in vitro and in vivo. Mge1 acts as a switch for sensing oxidative stress within the cell by changing its conformation from an active dimer to an inactive monomer. This conformational change in Mge1 is translated into a reduced interaction of Mge1 with mHsp70, thereby affecting the function of mHsp70 that is required for the vectorial transport of proteins across the mitochondrial cell membranes (Figure 7).
FIGURE 7:
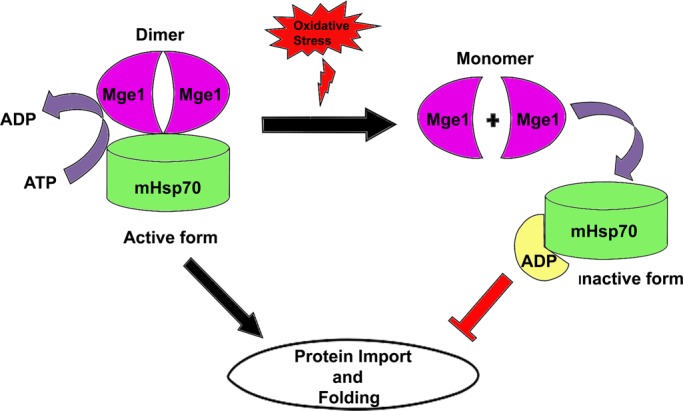
Schematic representation of effect of oxidative stress on Mge1-mediated regulation of the mitochondrial Hsp70 ATPase cycle.
On the basis of our results, we speculate that during oxidative stress, mHsp70-ADP remains associated with precursor protein that is being translocated across the inner membrane of mitochondria. Our studies show that there is reduced interaction of Mge1 with mHsp70 during oxidative stress, and this might be reflected in decreased ADP-to-ATP exchange on mHsp70. As a result, mitochondria might be stalling the transport of proteins into the mitochondrial matrix to prevent accumulation of unfolded proteins in the mitochondria. Our ATPase assays (Figure 6B) indicate that Mge1-M155L is efficient in stimulating ATPase activity of mHsp70 in the presence of H2O2. In addition, import of mitochondrial proteins and growth of yeast cells are not significantly reduced in the presence of Mge1-M155L mutant and H2O2 (Figure 6A). This persistent active Mge1-M155L during oxidative stress may be harmful to cells, as continuous import of unfolded proteins would cause protein aggregation in mitochondria. However, Mge1 plays a role in folding and degradation of misfolded proteins by interacting with chaperones (Westermann et al., 1995). It is possible that the mutant enhances the folding capability of Hsp70-family chaperones and may prevent the damage of mitochondria, thereby conferring a growth advantage. However, in the case of the wild-type strain, the import of precursors into mitochondria is defective, and cells might die due to the unavailability of proteins to elicit an adoptive response toward stress.
Using yeast as a model system, we showed that generation of ROS in mitochondria by the addition of antimycin A destabilizes Mge1–mHsp70 complex formation (Figure 5). To support this theory, we observed that the processing of matrix-targeted precursor protein is not altered with ROS induction. However, there is a delay in protease protection of the imported protein (Figure 6). This could be due to reduced interaction of Mge1 with mHsp70 and could lead to a delay in the release of substrate protein into the matrix, as ADP-bound Hsp70 molecules have higher affinity for substrate protein (Schneider et al., 1996). The precursor protein is arrested in the import channel and becomes susceptible to externally added protease. However, Mge1-M155L mutant does not reduce its interaction with Hsp70 in the presence of oxidant and functions as a nucleotide exchange factor to exchange ATP for ADP on Hsp70 to reinitiate the Hsp70 cycle. Mge1 is also required for Ssq, a second mHsp70 localized in mitochondria, which is required for import and assembly of Fe–S proteins (Knight et al., 1998). Intriguingly, Δssq cells are sensitive to growth on H2O2 plates. Although overexpression of Ssc (mHsp70) in Δssq cells suppresses some of the defects caused by ssq deletion, it does not rescue the slow growth on H2O2 plates (Voisine et al., 2000). This effect may be due to alteration in the structure of Mge1 and reduced interactions with mHsp70 and Ssq under these conditions. Further, we observed three or four high–molecular weight complexes when we performed in organelle cross-linking and probed with Mge1 (Figure 5B), but we could not pull down these complexes, possibly due to epitope specificity (Figure 5, B and C). These additional complexes that we observed in organelle cross-linking are likely to be Ssq-Mge1–containing complexes or other Mge1-containing complexes.
Methionine, cysteine, tryptophan, histidine, and tyrosine are prone to modifications during oxidative stress (Kim et al., 2001). In the presence of oxidative stress these amino acids are modified by large polar sulfoxide groups that increase the affinity for hydrogen bonding, leading to destabilization of the proteins. It is assumed that methionine serves as a ROS scavenger in the cell to protect cellular structure and integrity (Levine et al., 1999). Of interest, yeast Mge1 protein lacks cysteines and contains only one methionine. Mutating the lone methionine to leucine shows the effect on Mge1 regulation during oxidative stress both in vitro and in vivo (Figures 1, 2, and 4–6).
In summary, we provided clear evidence that Mge1 responds to oxidative stress and its activity depends on the metabolic and redox environment of the matrix. Our results also suggest that Mge1 plays a key role in regulating the mitochondrial Hsp70 chaperone system and thereby mitochondrial metabolism during oxidative stress. This mechanism of Mge1 action during oxidative stress appears to be conserved across evolution from yeast to humans. It is likely that there are other stresses besides heat and oxidative stress that might elicit the same response from Mge1.
MATERIALS AND METHODS
Plasmid construction
The complete coding sequence of yeast MGE1 was amplified without the mitochondrial targeting sequence (Δ43) by using forward primer MGE1_Fwd1 (5′-CCCAGAATTCACCATGGATGAAGCCAAAAGTGAAGAATCC-3′) and reverse primer MGE1_Rev1 (5′-ATTCTCGAGTTAGTTCTCTTCGCCCTTAACAATTCC-3′) and cloned into an E. coli expression vector pET28a+ to generate a hexahistidine tag at the N-terminal of MGE1 to generate pNB128. Human MGE1 without mitochondrial targeting sequence was amplified using HeLa cells cDNA as template, forward primer MGE1_Fwd2 (5′-CAAAGGATCCACCATGTCTCCCCGGTTGTTG-3′), and reverse primer MGE1-Rev2 (5′-ACCCCTCGAGCTAAGCTTCCTTCACCACCCC-3′) and cloned into pET28a+ vector to generate pNB182. Plasmids pNB184 harboring MGE1-M155L and MGE1-H130L, respectively, were created by site-directed mutagenesis of MGE1 in pNB128. Wild-type MGE1 with presequence was amplified by using yeast genomic DNA using forward primers MGE1_Fwd3 (5′-CCCAGAATTCACCATGAGAGCTTTTTCAGCAGCC-3′) and reverse primer MGE1_Rev3 (5′-ATTCTCGAGTTAGTTCTCTTCGCCCTTAACAATTCC-3′). The PCR product was digested with EcoRI/XhoI restriction enzymes and cloned into high-copy URA3 and high-copy LEU2 plasmids under TEF promoters at similar restriction sites to generate pNB185 and pNB186, respectively. Site-directed mutagenesis was used to create pNB187 and pNB188 harboring MGE1-M155L and MGE1-M155L, respectively, from pNB186. Mitochondrial HSP70 plasmid in pet21b vector (pNB200) was a kind gift from Debkumar Pain (UMDNJ–New Jersey Medical School, Newark, NJ).
Yeast strains
A heterozygous diploid strain of MGE1, yNB39 (European Saccharomyces cerevisiae Archive for Functional Analysis [EUROSCARF], Institute for Molecular Biosciences, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany) (Mat a/alpha:hisΔ1/3his3Δ1;Leu2Δ0/leu2Δ0;lys2Δ0/LYS2;MET15/met15Δ0;ura3Δ0/ura3Δ0;OR232w::kanMX4/YOR232w), a derivative of BY4743, was used for making haploid strains expressing either wild type or MGE1 mutants for in vivo studies. Briefly, yeast strain yNB39 was transformed with pNB185 to generate strain yNB59. yNB59 strain was grown in 2% KOAc with minimal amino acids to induce sporulation. Random sporulated colonies were selected for haploid yNB65 by markers present on the genetic background. Haploid strain yNB65B was confirmed by PCR (primer sequence from the EUROSCARF website).
The high-copy URA3 plasmid in the yNB65 strain was replaced with either pNB186 or pNB188 or pNB187 by plasmid shuffling to generate yNB71, yNB72, or yNB73 strains carrying wild-type MGE1 or MGE1-H130L or MGE1-M155L mutants, respectively. Briefly, yNB65 strain transformed with Leu plasmids was treated with 5-fluoroorotic acid for two generations to remove URA3-borne wild-type copy, and the loss of the URA3 plasmid was confirmed by lack of growth on SC-Ura plate. The resulting strains yNB71 (MGE1 wild type), yNB72 (MGE1-H130L), and yNB73 (MGE1-M155L) were used for further analysis.
Yeast media
Standard yeast protocols were used for culturing yeast strains. Wild-type yeast strains were grown in 1% yeast extract, 2% peptone, and 2% dextrose (YPD). Synthetic dextrose (SD) minimal medium contained 0.73% yeast nitrogen base with amino acids, 0.4% ammonium sulfate, and 2% glucose. For mitochondria isolation, we used semisynthetic lactose medium containing 2% lactose with yeast extract, ammonium chloride, calcium chloride, magnesium chloride, potassium phosphate, and glucose as described (Sepuri et al., 1998b). Yeast cells were grown at 30°C. Yeast transformations were performed by following the standard lithium acetate method (Gietz and Woods, 2006).
Purification of recombinant proteins
E. coli BL21 (DE3) Codon Plus (RIL) cells were transformed with pNB128, pNB182, pNB183, and pNB200 plasmids carrying wild-type MGE1, MGE1-M155L, human MGE1, and mhsp70, respectively. Expression of proteins was induced in the presence of 1 mM isopropyl-1-thio-β-d-galactopyranoside. The soluble histidine-tagged, wild-type, mutant yeast Mge1, human Mge1, and mHsp70 proteins were purified using Talon metal affinity resin (Invitrogen, Carlsbad, CA) and concentrated. Based on silver staining, wild type and mutant Mge1 constituted ∼95% of the total purified protein, with a minor DnaK contaminant. Yeast mHsp70 purified as a single protein. Antibodies against yeast Mge1 were raised in rabbit, and monospecific antibodies were purified by using CNBr antigen–coupled Sepharose beads. Purified GST protein was a kind gift from Bramanandam Manavathi (University of Hyderabad, Hyderabad, India).
Preparation of H2O2 solution
Initially a stock of 1 M H2O2 was prepared from commercially available 30% (vol/vol) H2O2 (Merck, Darmstadt, Germany) by taking 102.14 μl of H2O2 and diluting it to 1 ml with deionized water. The concentration of H2O2 was measured spectrophotometrically at OD240 by placing 1 ml of a 10 mM solution in a quartz cuvette. The concentration of H2O2 was calculated by using its extinction coefficient value (E240 = 43.6 M−1 cm−1).
In vitro cross-linking
Cross-linking of purified yeast wild-type Mge1, Mge1-M155L mutant, human Mge1. and GST protein was performed with BS3 (Pierce, Indianapolis, IN) at room temperature in 20 mM sodium phosphate (pH 7.4), 150 mM NaCl, and 50 mM borate. We preincubated 2.5 μg of purified proteins with increasing concentrations of H2O2 (0.5, 1, and 2 mM) at room temperature for 20 min and added BS3 to a final concentration of 0.1 mM. The reaction was quenched by addition of 40 mM Tris-HCl (pH 7.5) and analyzed by SDS–PAGE.
In organelle cross-linking and immunoprecipitation
Mitochondria were isolated from yeast as described (Sepuri et al., 2007, 2012). Mitochondria isolated from yNB71 and yNB73 were preincubated with increasing concentrations of antimycin A on ice for 20 min in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.4), followed by addition of the membrane-permeable cross-linker disuccinimidyl suberate (DSS; Pierce) at 100 μM final concentration. The reaction was incubated for 30 min at room temperature and quenched by addition of 40 mM Tris-HCl (pH 7.5). Samples were either analyzed directly by SDS–PAGE or subjected to immunoprecipitation.
Mitochondria were solubilized in lysis buffer (50 mM Tris-HCl. pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors) and incubated on ice for 30 min. Lysates were centrifuged for 15 min at 13,000 rpm at 4ºC. The clear supernatant fractions were incubated with protein A beads for 1 h at 4ºC to remove nonspecific interactions. Precleared lysates were incubated with Mge1 antibody for 5 h at 4ºC on a rotator. Protein A beads were added to the lysates and further incubated for another 4 h at 4ºC. The beads were washed four times with mitochondrial lysis buffer. Samples were resolved on SDS–PAGE, Western transferred, and probed with antibodies specific to mHsp70.
Circular dichroism spectroscopy
Circular dichroism spectra and oxidative denaturation curves were obtained in a Jasco J-810 spectropolarimeter using a 2-mm-path-length cuvette. Samples were dialyzed against 20 mM sodium phosphate (pH 7.0) overnight. Four spectra were recorded between 260 and 190 nm and averaged. The secondary structure was determined using CDNN 2.1 software for protein secondary structure analysis.
Measurement of ROS
Yeast strains yNB71 and yNB73 carrying wild-type Mge1 and Mge1-M155L were grown overnight in YPD medium. Cells (107 cells/ml) were preincubated in YPD medium with 40 μM dichlorofluorescin diacetate at 30°C for 1 h, followed by incubation with increasing concentrations of antimycin A at 30°C for 1 h. Then the cells were isolated by centrifugation and were washed with phosphate-buffered saline. The resulting cell pellet was resuspended in 100 μl of phosphate-buffered saline. Fluorescence intensity of the cell suspension containing 107 cells was read with a fluorescence spectrophotometer (PerkinElmer, Waltham, MA) at an excitation wavelength of 480 nm and an emission wavelength of 525 nm (Ryu et al., 2003).
In vitro protein import.
Radiolabeled precursor proteins were generated by in vitro transcription/translation in the presence of [35S]methionine by using rabbit reticulocyte lysate (Promega, Madison, WI; Tammineni et al., 2012). Mitochondria from Mge1 wild type and M155L mutant were treated with either 20 μM antimycin A or 5 μM valinomycin on ice for 15 min. The mitochondrial samples were incubated with import buffer (fatty acid–free bovine serum albumin, sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM methionine, 10 mM 3-(N-morpholino)propanesulfonic acid/KOH, pH 7.0, 5 mM ATP, pH 7.0, and 1 mM dithiothreitol) with [35S]methionine-labeled precursor proteins at 30°C for different time points. After the indicated times, mitochondria were isolated by centrifugation and washed in SEM (250 mM sucrose, 1 mM EDTA, 20 mM MOPS/KOH, pH 7.0) buffer. The samples were either directly subjected to SDS–PAGE or protease protection analysis in the presence of trypsin for 15 on ice, followed by inhibiting the trypsin by soybean trypsin inhibitor and washing the mitochondria samples in SEM buffer. The samples were separated on SDS–PAGE and analyzed by autoradiogram.
ATPase assay of mHsp70.
The ATPase activity of mhsp70 was analyzed by measuring the release of Pi from [γ-32P]ATP using the method as described (Viitanen et al., 1990). Recombinant mHsp70 (1 μg) was incubated with or without 5 μg of wild type or Mge1-M155L mutant in an ATPase assay buffer (50 mM HEPES/KOH, pH 7.2, 5 mM MgCl2, and 100 mM KCl) in the presence of 0.05 mM [γ-32P]ATP (3000 Ci/mmol). At different time intervals 25 μl of reaction mixture was taken and mixed with seven volumes of 1 M perchloric acid and 1 mM sodium phosphate and kept on ice. After addition of 16 volumes of 20 mM ammonium molybdate and isopropyl acetate, the samples were mixed and centrifuged at 15,000 × g for 30 s in a microfuge to separate organic and inorganic phases. The amount of radioactive Pi released as phosphomolybdate complex in organic phase was measured by scintillation counting. Control reactions were performed to eliminate the background nonspecific hydrolysis of ATP.
Supplementary Material
Acknowledgments
This work was supported by grants from the Council of Scientific and Industrial Research, India (38(1194)08/EMR-II), to N.B.V.S. and the Department of Science and Technology/Promotion of University Research and Scientific Excellence, Department of Science and Technology/Funds for Improvement of Infrastructure for Science and Technology, Department of Biotechnology/Centre for Research and Education in Biology, and Biotechnology and University Grants Commission/Centre of Advanced Study funds to the School of Life Sciences and Department of Biochemistry, University of Hyderabad. We thank Debkumar Pain for providing us with mHsp70 plasmid and mHsp70 antibodies. We thank Rajagopal Subramanyam, Hyderabad Central University, for helping us in analyzing the CD spectra data. We thank all the members of the N.B.V.S. laboratory and Thanuja Krishnamoorthy for comments and suggestions.
Abbreviations used:
- Ccpo
cytochrome c peroxidase
- DHFR
dihydrofolate reductase
- HSP 70
heat shock protein 70
- ROS
reactive oxygen species
- Tim
translocase of inner membrane
- Tom
translocase of outer membrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0719) on January 23, 2013.
REFERENCES
- Azem A, Oppliger W, Lustig A, Jeno P, Feifel B, Schatz G, Horst M. The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J Biol Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocheme HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Schmidt E, Koehler CM. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J Cell Biol. 2002;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiae’s Mge1p, the mitochondrial cochaperone of Ssc1p. J Biol Chem. 1996;271:23960–23966. doi: 10.1074/jbc.271.39.23960. [DOI] [PubMed] [Google Scholar]
- Demasi AP, Pereira GA, Netto LE. Yeast oxidative stress response. Influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. FEBS J. 2006;273:805–816. doi: 10.1111/j.1742-4658.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- Drechsel DA, Patel M. Differential contribution of the mitochondrial respiratory chain complexes to reactive oxygen species production by redox cycling agents implicated in parkinsonism. Toxicol Sci. 2009;112:427–434. doi: 10.1093/toxsci/kfp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz R, Marszalek J, Schilke B, Craig EA, Lill R, Muhlenhoff U. The Hsp70 chaperone Ssq1p is dispensable for iron-sulfur cluster formation on the scaffold protein Isu1p. J Biol Chem. 2006;281:7801–7808. doi: 10.1074/jbc.M513301200. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc Natl Acad Sci USA. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- Grimshaw JP, Jelesarov I, Schonfeld HJ, Christen P. Reversible thermal transition in GrpE, the nucleotide exchange factor of the DnaK heat-shock system. J Biol Chem. 2001;276:6098–6104. doi: 10.1074/jbc.M009290200. [DOI] [PubMed] [Google Scholar]
- Grimshaw JP, Jelesarov I, Siegenthaler RK, Christen P. Thermosensor action of GrpE. The DnaK chaperone system at heat shock temperatures. J Biol Chem. 2003;278:19048–19053. doi: 10.1074/jbc.M300924200. [DOI] [PubMed] [Google Scholar]
- Groemping Y, Reinstein J. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol. 2001;314:167–178. doi: 10.1006/jmbi.2001.5116. [DOI] [PubMed] [Google Scholar]
- Kim YH, Berry AH, Spencer DS, Stites WE. Comparing the effect on protein stability of methionine oxidation versus mutagenesis: steps toward engineering oxidative resistance in proteins. Protein Eng. 2001;14:343–347. doi: 10.1093/protein/14.5.343. [DOI] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- Kutik S, Guiard B, Meyer HE, Wiedemann N, Pfanner N. Cooperation of translocase complexes in mitochondrial protein import. J Cell Biol. 2007;179:585–591. doi: 10.1083/jcb.200708199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Dekker PJ, Voos W, Craig EA, Pfanner N. Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol Cell Biol. 1995;15:7098–7105. doi: 10.1128/mcb.15.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER. Methionine residues may protect proteins from critical oxidative damage. Mech Ageing Dev. 1999;107:323–332. doi: 10.1016/s0047-6374(98)00152-3. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Rizzuto R. Signaling pathways in mitochondrial dysfunction and aging. Mech Ageing Dev. 2010;131:536–543. doi: 10.1016/j.mad.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A, Pfanner N, Voos W. Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 2000;1:404–410. doi: 10.1093/embo-reports/kvd093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men L, Wang Y. The oxidation of yeast alcohol dehydrogenase-1 by hydrogen peroxide in vitro. J Proteome Res. 2007;6:216–225. doi: 10.1021/pr0603809. [DOI] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Berg A, Adam A, Neupert W, Hell K. Association of the Tim14.Tim16 subcomplex with the TIM23 translocase is crucial for function of the mitochondrial protein import motor. J Biol Chem. 2007;282:18037–18045. doi: 10.1074/jbc.M701895200. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro F, Muga A. Thermal adaptation of the yeast mitochondrial Hsp70 system is regulated by the reversible unfolding of its nucleotide exchange factor. J Mol Biol. 2006;358:1367–1377. doi: 10.1016/j.jmb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Brinker A, Paschen SA, Moarefi I, Hayer-Hartl M, Neupert W, Brunner M. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 2002;21:3659–3671. doi: 10.1093/emboj/cdf358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Lee Y, Han SK, Kim HY. The role of hydrogen peroxide produced by polychlorinated biphenyls in PMR1-deficient yeast cells. J Biochem. 2003;134:137–142. doi: 10.1093/jb/mvg121. [DOI] [PubMed] [Google Scholar]
- Sakuragi S, Liu Q, Craig E. Interaction between the nucleotide exchange factor Mge1 and the mitochondrial Hsp70 Ssc1. J Biol Chem. 1999;274:11275–11282. doi: 10.1074/jbc.274.16.11275. [DOI] [PubMed] [Google Scholar]
- Schiller D, Cheng YC, Liu Q, Walter W, Craig EA. Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol Cell Biol. 2008;28:4424–4433. doi: 10.1128/MCB.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HC, Westermann B, Neupert W, Brunner M. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 1996;15:5796–5803. [PMC free article] [PubMed] [Google Scholar]
- Schoneich C. Reactive oxygen species and biological aging: a mechanistic approach. Exp Gerontol. 1999;34:19–34. doi: 10.1016/s0531-5565(98)00066-7. [DOI] [PubMed] [Google Scholar]
- Schulke N, Sepuri NB, Gordon DM, Saxena S, Dancis A, Pain D. A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J Biol Chem. 1999;274:22847–22854. doi: 10.1074/jbc.274.32.22847. [DOI] [PubMed] [Google Scholar]
- Schulke N, Sepuri NB, Pain D. In vivo zippering of inner and outer mitochondrial membranes by a stable translocation intermediate. Proc Natl Acad Sci USA. 1997;94:7314–7319. doi: 10.1073/pnas.94.14.7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepuri NB, Gordon DM, Pain D. A GTP-dependent “push” is generally required for efficient protein translocation across the mitochondrial inner membrane into the matrix. J Biol Chem. 1998a;273:20941–20950. doi: 10.1074/jbc.273.33.20941. [DOI] [PubMed] [Google Scholar]
- Sepuri NB, Gorla M, King MP. Mitochondrial lysyl-tRNA synthetase independent import of tRNA lysine into yeast mitochondria. PLoS One. 2012;7:e35321. doi: 10.1371/journal.pone.0035321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepuri NB, Schulke N, Pain D. GTP hydrolysis is essential for protein import into the mitochondrial matrix. J Biol Chem. 1998b;273:1420–1424. doi: 10.1074/jbc.273.3.1420. [DOI] [PubMed] [Google Scholar]
- Sepuri NB, Yadav S, Anandatheerthavarada HK, Avadhani NG. Mitochondrial targeting of intact CYP2B1 and CYP2E1 and N-terminal truncated CYP1A1 proteins in Saccharomyces cerevisiae—role of protein kinase A in the mitochondrial targeting of CYP2E1. FEBS J. 2007;274:4615–4630. doi: 10.1111/j.1742-4658.2007.05990.x. [DOI] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16:3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci USA. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem. 2012;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Neupert W, Cyr DM. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Lubben TH, Reed J, Goloubinoff P, O'Keefe DP, Lorimer GH. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- Voisine C, Schilke B, Ohlson M, Beinert H, Marszalek J, Craig EA. Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol. 2000;20:3677–3684. doi: 10.1128/mcb.20.10.3677-3684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Prip-Buus C, Neupert W, Schwarz E. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 1995;14:3452–3460. doi: 10.1002/j.1460-2075.1995.tb07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Reichenbecher V. The effects of superoxide and the peripheral benzodiazepine receptor ligands on the mitochondrial processing of manganese-dependent superoxide dismutase. Exp Cell Res. 1999;246:443–450. doi: 10.1006/excr.1998.4331. [DOI] [PubMed] [Google Scholar]
- Wright G, Reichenbecher V, Green T, Wright GL, Wang S. Paraquat inhibits the processing of human manganese-dependent superoxide dismutase by SF-9 insect cell mitochondria. Exp Cell Res. 1997;234:78–84. doi: 10.1006/excr.1997.3579. [DOI] [PubMed] [Google Scholar]
- Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin A, Gabrielli N, Calvo IA, Garcia-Santamarina S, Hoe KL, Kim DU, Park HO, Hayles J, Ayte J, Hidalgo E. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS One. 2008;3:e2842. doi: 10.1371/journal.pone.0002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.