Table 3. Biclusters divided by kind of infection.
| Pathogens | Bicluster 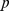 -value -value |
# Gene Sets | # Targets | Target Enrich. (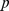 -value) -value) |
| Gastrointestinal | ||||
| Yersinia Enterocolitica wap and p60 strains, Helicobacter Pylori, and Escherichia Coli |
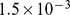
|
227 | 18 |
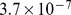
|
| Yersinia Enterocolitica, Lactobacillus Acidophilus, Listeria Monocytogenes, and Helicobacter Pylori |
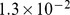
|
173 | 11 |
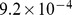
|
| Yersinia Enterocolitica and Helicobacter Pylori |
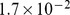
|
272 | 21 |
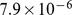
|
| Yersinia Enterocolitica, Listeria Monocytogenes, and Bifidobacterium Bifidum |
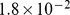
|
269 | 17 |
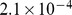
|
| Yersinia Enterocolitica, Bifidobacterium Bifidum, Streptococcus Pyogenes, and Helicobacter Pylori |
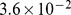
|
101 | 6 |
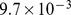
|
| Respiratory | ||||
| Pseudomonas Aeruginosa, and Mycobacterium Tuberculosis |
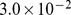
|
245 | 16 |
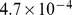
|
| Hematopoietic | ||||
| Ehrlichia Chaffeensis; Strains: arkansa and wakulla |
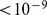
|
979 | 186 |
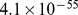
|
The table shows the biclusters that contained pathogens that cause an infection in a single type of tissue. The columns from left to right are: (i) list of pathogens contained in a bicluster, (ii) a 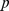 -value indicating the statistical significance of the bicluster, (iii) the number of gene sets in the bicluster, (iv) the number of known human drug target genes/proteins in the bicluster, and (v)
-value indicating the statistical significance of the bicluster, (iii) the number of gene sets in the bicluster, (iv) the number of known human drug target genes/proteins in the bicluster, and (v) 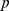 -value indicating the enrichment of the bicluster in know human drug-target genes/proteins.
-value indicating the enrichment of the bicluster in know human drug-target genes/proteins.
