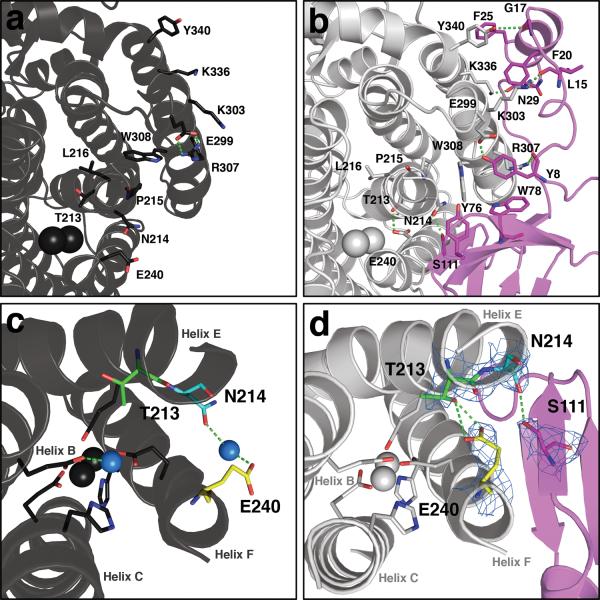
Figure 2. Conformational changes near the diiron center and pore residues in MMOH upon MMOB binding.
In both (a) and (b), protein backbones are shown as ribbons in black (MMOHox), grey (MMOH from the H-B complex), and magenta (MMOB from the H-B complex). Iron atoms are depicted as black (MMOHox) or grey (H-B) van der Waals spheres. Interactions between key residues at the protein-protein interface are depicted as sticks; carbon atoms are colored to match to the protein backbone from which they stem, nitrogen atoms are shown in blue, and oxygen atoms in red; hydrogen bonds are represented as green dashes. Conformational changes in the MMOH pore residues upon MMOB binding are illustrated in MMOHox (c) and H-B (d). Iron-ligating helices B, C, E, and F and iron atoms are shown as black (c) and grey (d) ribbons and MMOB as magenta ribbons. Active site side chain ligands are shown as sticks in grey. Residues Thr 213 (green), Asn 214 (cyan), and Glu 240 (yellow) in MMOH, and Ser 111 in MMOB (magenta), are rendered as sticks. Nitrogen and oxygen atoms are shown in blue and red, respectively. The blue spheres in (c) are water molecules or hydronium ion. 2Fo-Fc electron density at 1.0 sigma from the H-B structure is drawn as a light blue mesh about key residues. The Glu 240 carboxylate side chain may function to deliver protons from solvent to the diiron site while closing the pore (see text).