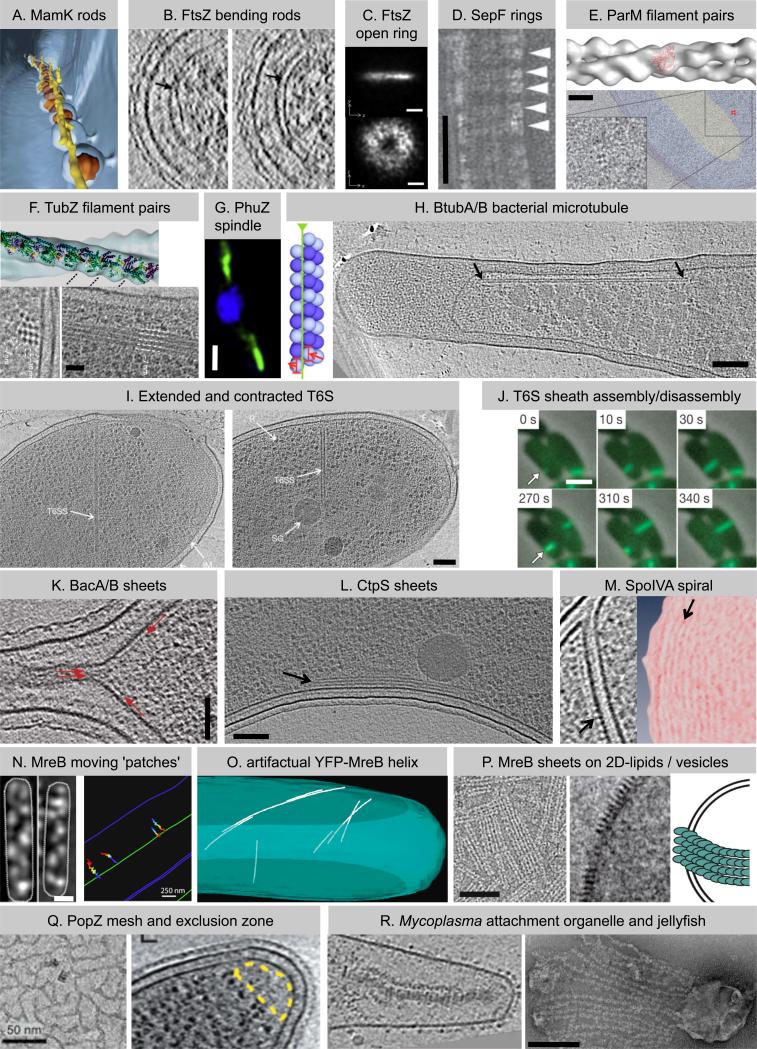
Figure 1. Superstructures of the bacterial cytoskeleton.
(A) Individual MamK rods (yellow in shown segmentation) organize the magnetosome chain [7]. (B) ECT of wildtype cells identified the cell division protein FtsZ (arrows) as straight (left) or bent (right) rods [13]. (C) Superresolution fLM observed FtsZ as a ring-shaped signal with uneven density (y, cell length axis) [19]. (D) SepF bundles FtsZ filaments (running vertical) in vitro by forming rings (arrowheads) around them [25]. (E) Plasmid segregating ParM forms twisted filament pairs in vitro (upper [30]) and small bundles of filament pairs in vivo (lower: perpendicular view [33]). (F) TubZ also segregates plasmids, assembles twisted filament pairs in vitro (upper) and bundles in vivo (lower: perpendicular (left) and longitudinal (right) view of overexpressed TubZ) [42]. (G) PhuZ forms a spindle-like structure (green) that positions phage DNA at midcell (blue, phage/bacterial DNA) [44]. (H) BtubA/B assemble bacterial microtubules (left: model) which localize close to the cytoplasmic membrane in vivo (right, arrows) [50]. (I) Type VI secretion (T6S) requires a contractile phage tail-like sheath which is found in extended (left) and contracted (right) confirmation [59]. (J) The tubular T6S sheath is dynamic and cycles between assembly (upper row), quick contraction, and disassembly (lower row) [59]. (K) Bactofilins [65] and (L) CTP synthase [67] both form sheets (arrows) at the cytoplasmic membrane, where they recruit other proteins. (M) Densities thought to represent SpoIVA (left, arrow) form concentric rings (right panel shows density projections, arrow) on the mother side of the outer spore membrane, where SpoIVA recruits spore coat proteins [79]. (N) MreB forms small patches moving circumferentially around the cell driven by the cell wall synthesis machinery (left: fLM images, right: traced patches) [87,88]. (O) Extended MreB helices in E. coli (segmentation shown) are an artifact of an N-terminal YFP tag [90]. (P) MreB forms filaments and sheets on 2D-lipids (left) and vesicles (middle and model on right) [91]. (Q) PopZ establishes a ribosome exclusion zone (right, yellow) in vivo presumably by the formation of a 3-D mesh (left, in vitro) [93,94]. (R) Gliding motility requires multi-element composites such as the attachment organelle in Mycoplasma pneumoniae (left, [96]) or the jellyfish-like multi-element complex in Mycoplasma mobile (right, [98]). Images are adapted with permissions from references listed in legend. Bars: 200nm in C, R; 50nm in D, K, Q; 100nm in E, H, I, L, P; 25nm in F; 1μm in G, J, N (left); 250nm in N (right).