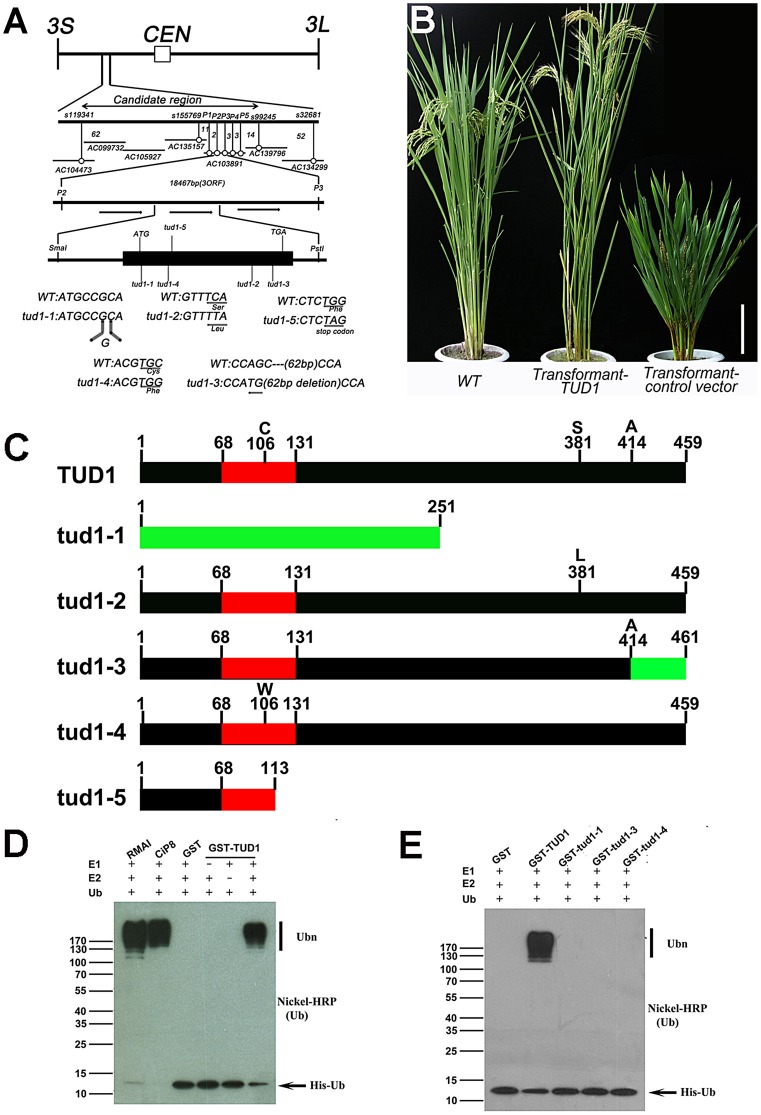
Figure 6. TUD1 is a functional E3 ligase.
(A) High resolution linkage and physical map of tud1 locus. Horizontal lines represent chromosome 3 and vertical bars the molecular markers. The numbers of recombinant plants are indicated between the markers. The physical map of the tud1 locus was constructed using 7 BAC clones and the candidate region of the tud1 mutation was found between markers P2 and P3. The genomic structure of the TUD1 gene and the positions of mutations in tud1 alleles are also shown. The black box indicates the single exon. The mutated DNA sequences of tud1 alleles are shown in the bottom. The tud1-1 has one-base insertion near the initiation codon. The tud1-2, tud1-4 and tud1-5 have different one-base substitutions in the coding sequence. The tud1-3 has a one-base substitution and 62 bp base deletion in the exon. CEN:centromere, 3S: the short arm of chromosome 3, 3L: the long arm of chromosome 3. (B) Phenotypic complementation by introduction of the TUD1 gene. Nipponbare was the parent for tud1-2 and used as the wild-type plant. Left, the wild type (Nipponbare); center, transformant-TUD1; right, transformant-control vector. Bar: 10 cm. (C) Schematic structures of TUD1 protein and its mutant alleles. Black bars indicate TUD1 coding regions. Red bars depict U-box motifs. Green bars represent the aberrant truncated protein of tud1-1 and partial amino acids residues of tud1-3 due to a frameshift mutation. Numbers represent the amino acids of protein sequences. (D) Ubiquitination assays with GST-TUD1. GST-TUD1, GST, and positive controls of RMAI and CiP8 were assayed for E3 activity in the presence of E1 (from wheat), E2 (UBCh5b), and 6×His tag ubiquitin (Ub). The numbers at left denote the molecular mass in kilodaltons. Samples were resolved by 8% SDS-PAGE. The nickel-horseradish peroxidase was used to detect His tag ubiquitin. (E) Ubiquitination assays with GST-TUD1 and its mutant variants. GST-TUD1 and its mutant forms (GST-tud1-1, GST-tud1-3 and GST-tud1-4 fusion proteins) were assayed for E3 activity. The reaction conditions were the same as in (D).