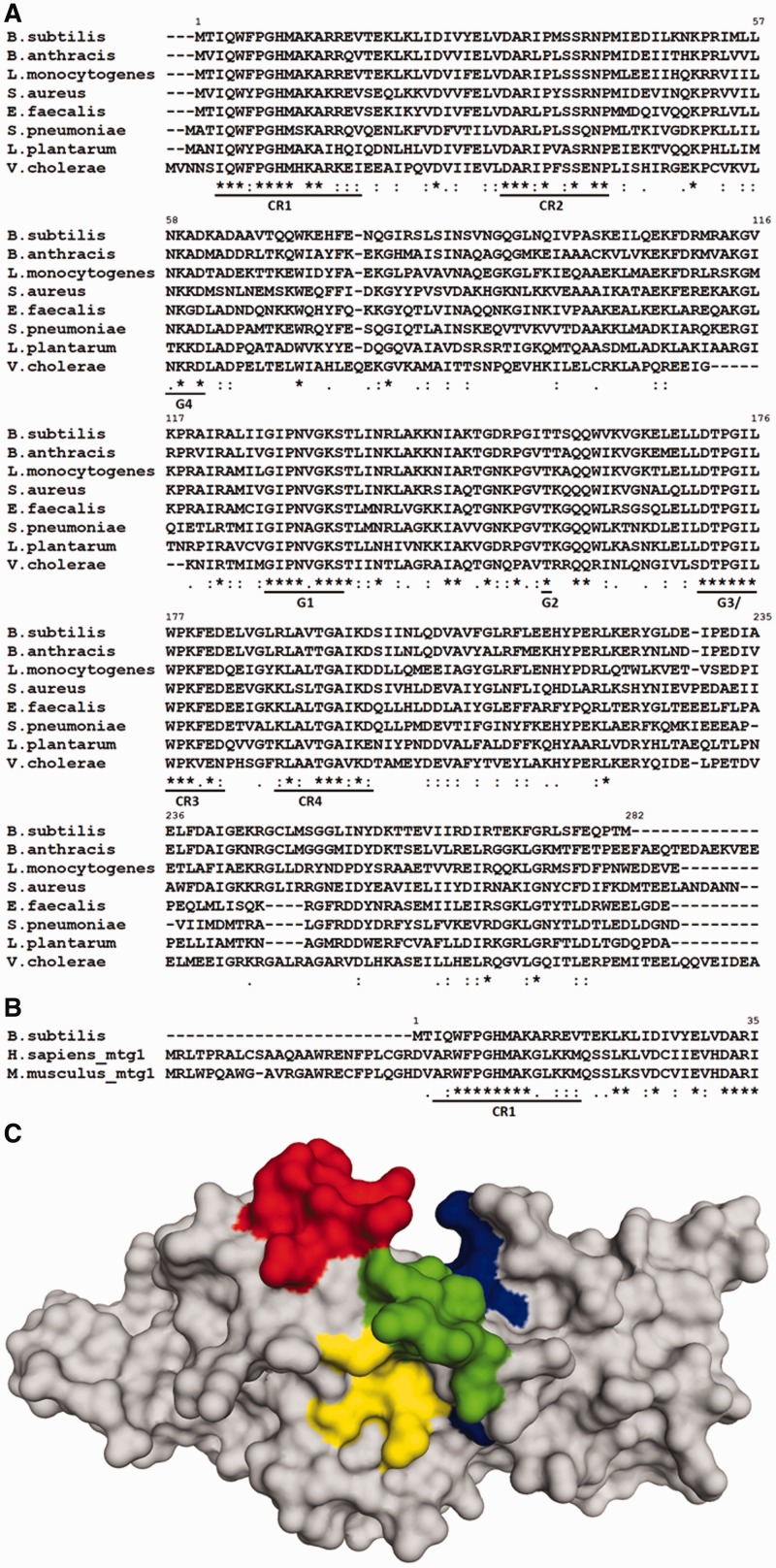
Figure 1.
(A) Multiple sequence alignment of selected bacterial RbgA homologs. Highly conserved regions are underlined. G-domain motifs are indicated as G4 (NKXD), G1 (GXXXXGKS/T), G2 (T) and G3 (DXXG) in addition to four conserved regions (CR1-CR4). (B) Multiple sequence alignment of RbgA with eukaryotic homologs Mtg1 from humans and mouse highlighting conserved region CR1. Alignments were constructed with ClustalW with default parameters and species indicated to the left. ‘Asterisk’ indicates positions that have a single fully conserved residue; ‘colon’ indicates conservation between groups of strongly similar physicochemical properties; ‘dot’ indicates conservation between groups of weakly similar physicochemical properties. (C) Structure of RbgA depicting CR1, CR2, CR3 and CR4. Crystal structure of RbgA (1PUJ) depicting the three conserved regions. First nine amino acid residues are not structured in the crystal. CR1 (9–17) is shown in red, CR2 (30–42) is shown in yellow, CR3 (175–182) is shown in green and CR4 (188–197) is shown in blue. All four conserved regions are dispersed across the protein sequence and lie in close vicinity in the structure of the protein. The amino acid numbering of RbgA from B. subtilis is indicated in each alignment.