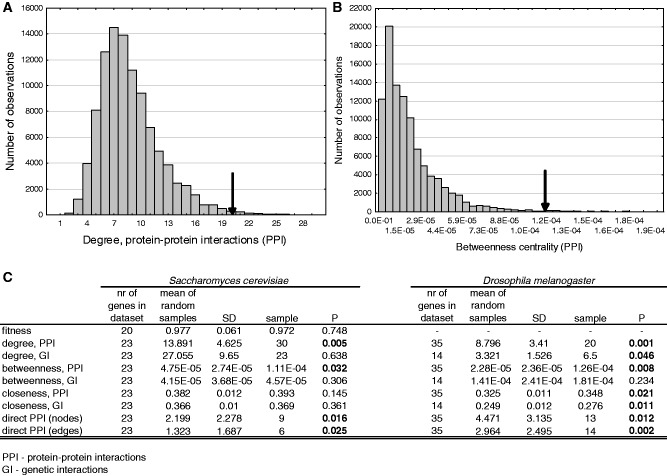
Figure 3.
Characteristics of the genes that were incorporated into TEs. We performed Monte Carlo simulations to test whether fitness and network characteristics like degree (the number of interactions with other nodes), betweenness centrality (the fraction of shortest paths that pass through a node) and closeness centrality (the inverse of the mean distance between node v and all other nodes reachable from it) are significantly different in the incorporated genes than the random expectation. (A) Distribution of the median degree (PPIs) in 100 000 random samples, the arrow represents the median of the 35 Drosophila genes for which PPI information was available. (B) Distribution of the median betweenness centrality in 100 000 random samples, and the observed level in Drosophila. (C) Statistical summary of Monte Carlo simulations. We did not perform tests for the fitness effect for Drosophila gene knockouts, as we are not aware of studies that provide such data at a genomic scale.