Figure 5.
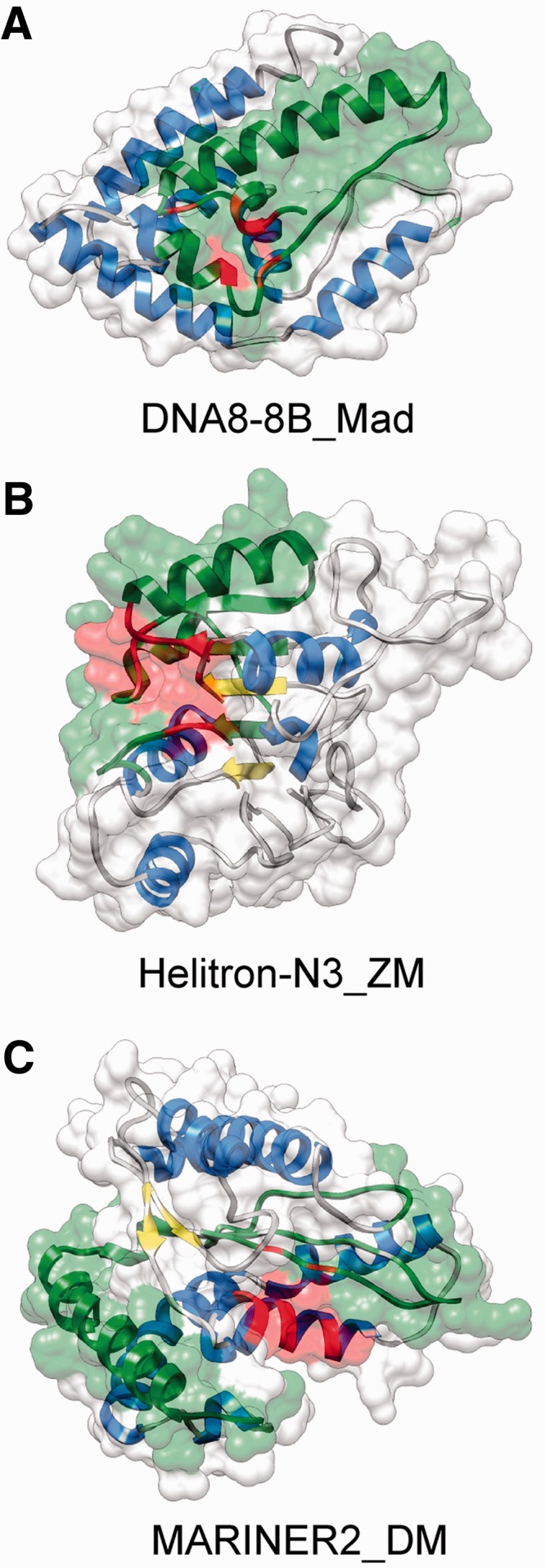
Examples of chimeric TE protein structures with an incorporated fragment of a host gene. The structures were predicted with I-TASSER, their function and catalytic centers were predicted with COFACTOR. In all cases, the estimated TM-score with the true topology is >0.5; thus, the models have an essentially correct topology. Alpha helices are highlighted with blue, beta sheets with yellow, green regions indicate the fraction of the sequence that is homologous to a non-TE protein and red highlights the catalytic core predicted by COFACTOR. (A) DNA8-8B_Mad from the apple genome, estimated TM score with the correct fold is 0.53. The incorporated fragment shows 80% sequence similarity to a short chain dehydrogenase (B9RTW7) with oxidoreductase activity (GO:0016491). (B) Helitron_N3_ZM from maize, estimated TM score is 0.64. The incorporated fragment shows 90% amino acid sequence similarity to maize fibrillarin (B6T4G7). The predicted highest scoring gene ontology terms for the molecular function of the protein are methyltransferase activity (GO:0008168) and RNA binding (GO:0003723). (C) MARINER2_DM transposon from Drosophila, estimated TM score is 0.74. The incorporated fragment is only 30% similar to the Drosophila gene CG18367-PA. The highest scoring molecular function GO term is DNA binding (GO:0003677).
