Figure 3.
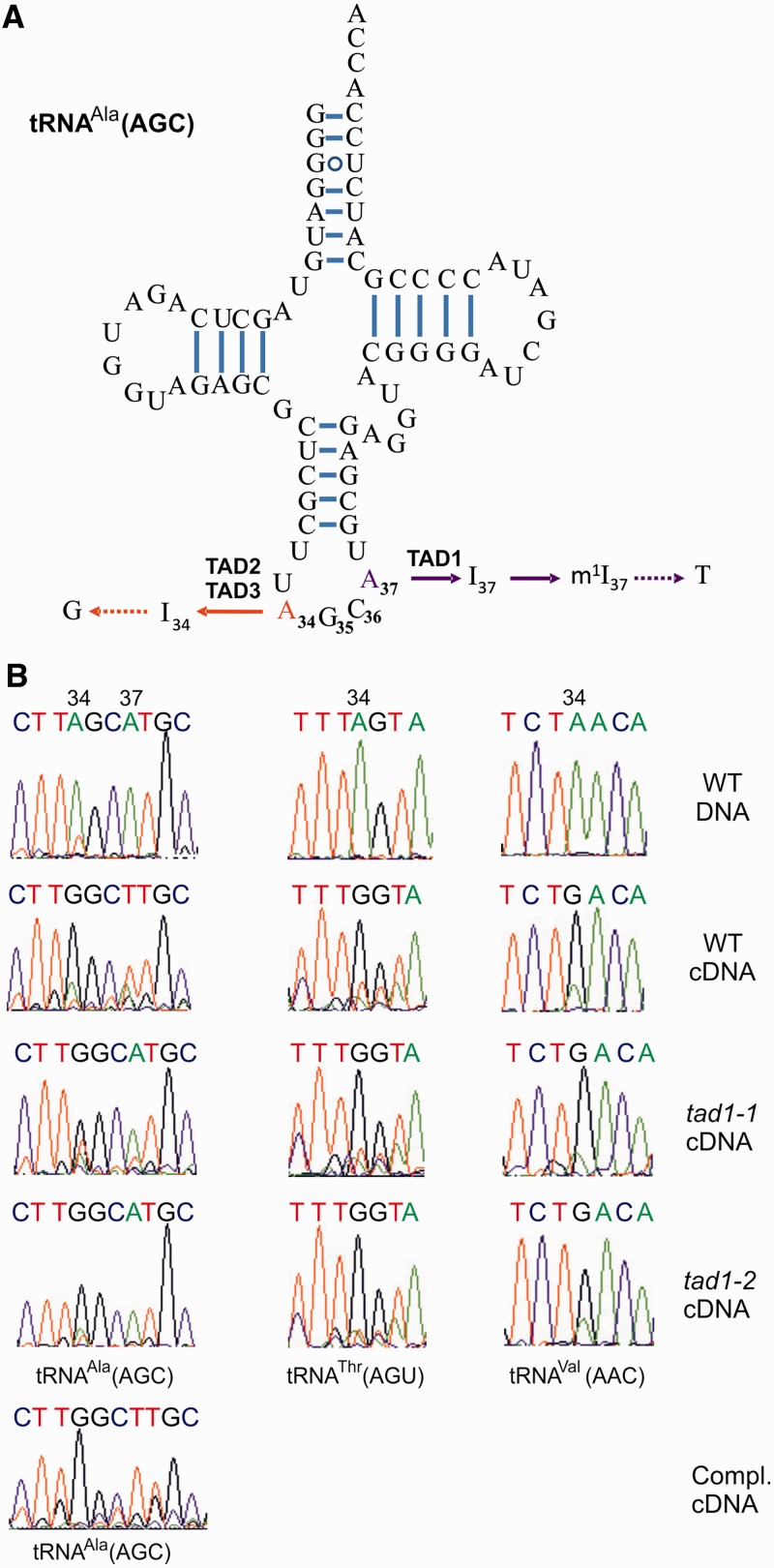
Analysis of tRNA-Ala(AGC) editing in tad1 mutants. (A) Cloverleaf structure of the tRNA-Ala(AGC) from Arabidopsis. Watson–Crick base pairing and UG base pairing are represented by bars and open circles, respectively. TAD1 deaminates A37 in the anticodon loop of the tRNA. I37 undergoes further modification by S-adenosylmethionine–dependent methylation to N1-methylinosine (m1I37; 45). m1I37 is read as T by reverse transcriptases (indicated by the dotted arrow). In yeast, two other deaminase proteins, TAD2 and TAD3, form a heterodimer and specifically deaminate A34 (46). Reverse transcriptases read I as G. (B) Analysis of A-to-I editing at positions 34 and 37 of tRNA-Ala(AGC) in wild-type Arabidopsis plants (WT), the tad1 mutants, tad1-1 and tad1-2, and the complemented tad1-2 line (Compl.). The tRNAs were reverse transcribed, the cDNA sequences amplified by PCR and directly sequenced. Note loss of A-to-I editing at position 37 in tad1 mutants (as evidenced by presence of the genomically encoded A instead of the T originating from reverse transcription of m1I-37 in the wild-type), but unaffected editing at position 34 (G indicating presence of inosine in both the wild-type and the mutants). As a control, two other inosine-containing cytosolic tRNAs (a threonine and a valine tRNA) were also investigated and, likewise, turned out to be unaffected in the Arabidopsis tad1 mutants. DNA indicates genomic DNA; cDNA denotes complementary DNA.