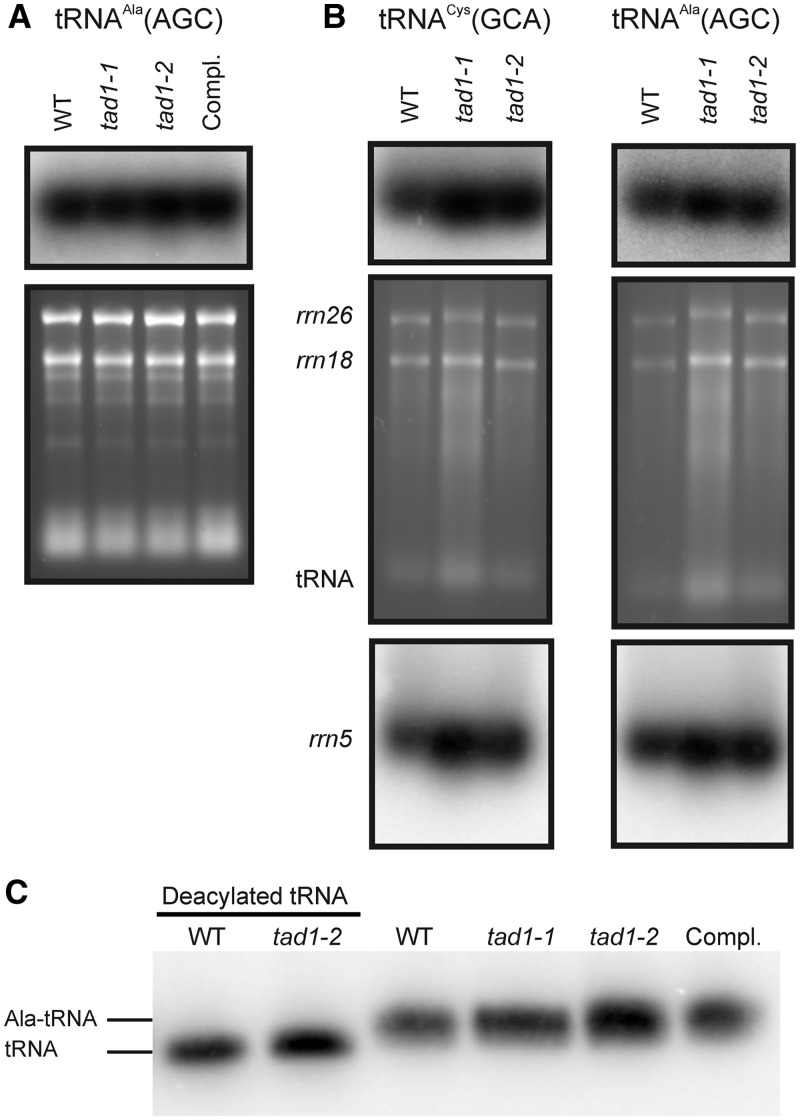
Figure 4.
Accumulation and aminoacylation of tRNA-Ala(AGC) in wild-type plants (WT), tad1 mutant plants and the complemented tad1-2 line (Compl.). Northern blot analyses were carried out with total cellular RNA (A) or purified mitochondrial RNA (B). To control for loading, the ethidium bromide-stained agarose gels before blotting are also shown. Accumulation of the mitochondrial encoded tRNA-Cys(GCA) was analyzed as a control in (B). The 26S and 18S rRNAs of the mitochondrial ribosome and the tRNA band are indicated in the ethidium bromide-stained agarose gels. To additionally control for loading differences visible in the mitochondrial RNA gel blots, the blots were stripped and re-hybridized to a probe specific to the mitochondrial 5S rRNA. (C) Analysis of aminoacylation of tRNA-Ala(AGC) in the wild-type, the tad1 mutant and the complemented tad1-2 line. In the gel system used here (42), the aminoacylated tRNA migrates slower than its corresponding deacylated species. To visualize the difference in electrophoretic mobility, aliquots of the wild-type sample and the tad1-2 sample were deacylated in vitro. Samples of 8 µg of RNA were separated by electrophoresis, blotted and hybridized to a tRNA-Ala(AGC)-specific probe.