Figure 3.
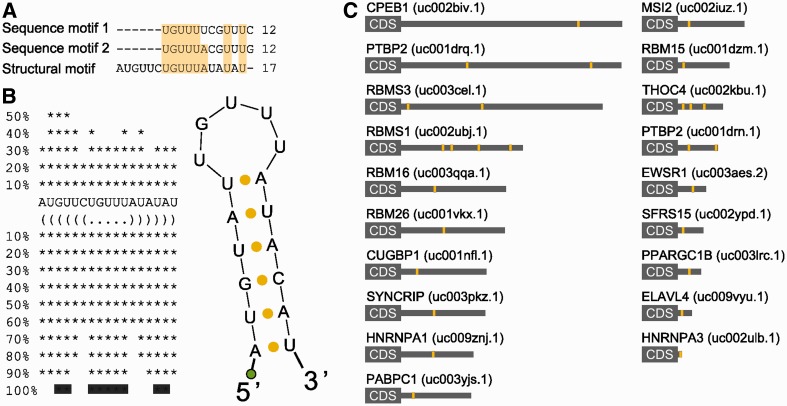
HCEs in mRNAs encoding RRM-type RBPs share a sequence and secondary structure motif. HCEs contained in the group of RRM-type RBP genes 3′-UTRs were scanned for both sequence and secondary structure motifs. The first search returned two, almost identical, 12-bases motifs; the second one produced a 17-bases hairpin which, after multiple alignment, emerged to contain a 12-bases core markedly similar to previously identified sequence motifs. This core represents the loop part of the hairpin which, as the two searches are quite concordant on it, may indeed represent a binding motif for the trans-factor of the regulatory network we are trying to identify. (A) shows the alignment between sequence and secondary structure motifs. Light-yellow background highlights column sequence identity. (B) shows the secondary structure motif and its bidimensional structure. The green circle represents the biotinylated DNA polyC linker added to the RNA. (C) Motif instances (yellow areas) mapped on the full length 3′-UTR (grey rectangle) of the 19 RRM-type RBP mRNAs. HGNC gene names are on the left, UCSC UTR names are on the right in parenthesis.