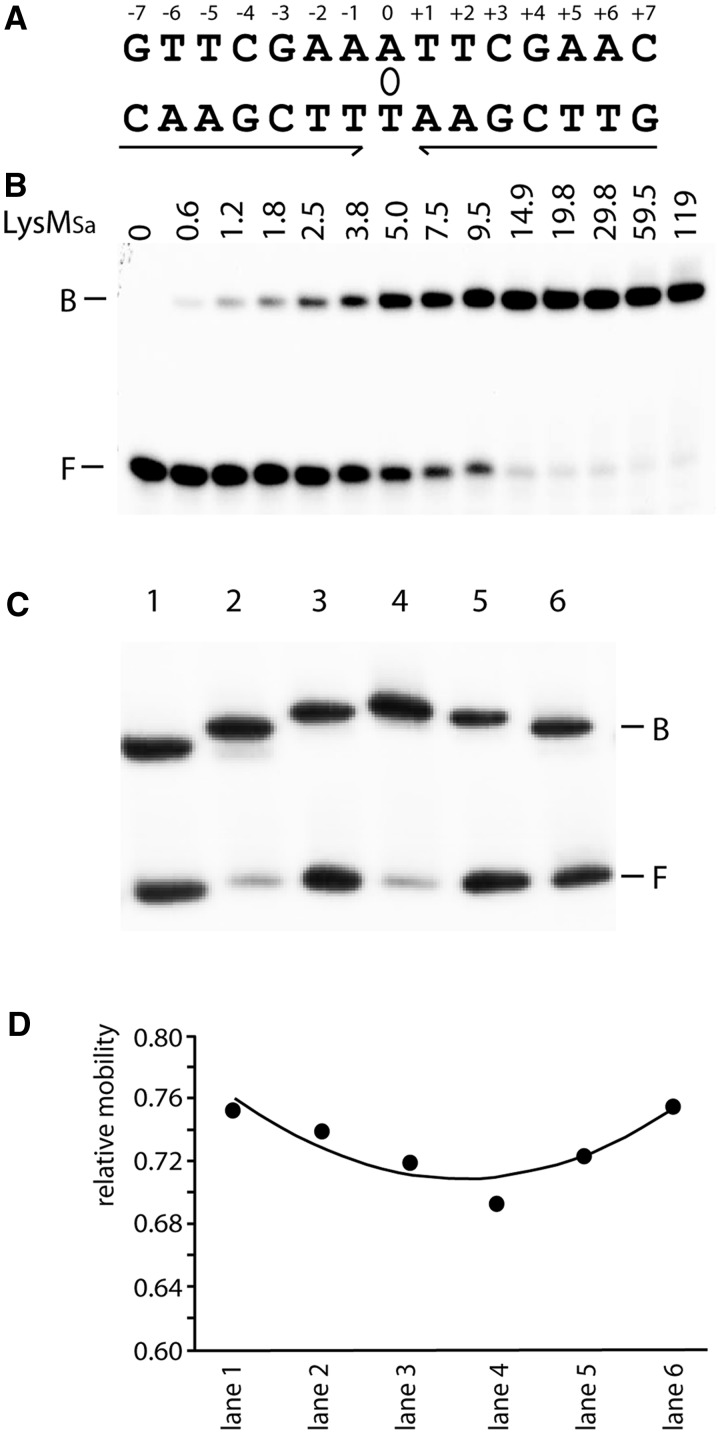
Figure 3.
Binding and LysMSa-induced DNA bending of the consensus binding site. (A) Cartoon representing the LysM consensus sequence. Inverted repeat elements are indicated with arrows; the axis of 2-fold symmetry is depicted by an ellipse. (B) EMSA of binding of LysMSa to a DNA fragment encompassing the consensus sequence. This fragment was generated by hybridization of 47-nt-long complementary oligonucleotides containing the 15-nt LysM box flanked on either side by the 16-nt stretches that surround the LysMSa binding site of the lysW operator in the S. acidocaldarius genome. Positions of bound (B) and free (F) DNA are indicated, as are applied LysMSa concentrations (in nM). (C) EMSA with permuted DNA fragments bearing the consensus binding site. In all binding reactions, we added 30 nM LysMSa. Characteristics of fragments are further described in Materials and Methods, and lane numbers in the EMSA correspond to fragment numbers. Positions of bound (B) and free (F) DNA are indicated. (D) Graphical representation of the relative mobility (μ) of different complexes as a function of the position of the insert sequence within the DNA fragment. The apparent bending angle (α) was calculated as follows: μM/μE = cos (α/2) (M = insert in the centre of the fragment, resulting in the lowest value of μ, and E = insert at the end of the fragment).